| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:27:18 UTC |
|---|
| Update Date | 2020-06-04 20:46:39 UTC |
|---|
| BMDB ID | BMDB0000244 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Riboflavin |
|---|
| Description | Riboflavin, also known as vitamin B2 or lactoflavin, belongs to the class of organic compounds known as flavins. Flavins are compounds containing a flavin (7,8-dimethyl-benzo[g]pteridine-2,4-dione) moiety, with a structure characterized by an isoalloaxzine tricyclic ring. Riboflavin is a drug which is used for the treatment of ariboflavinosis (vitamin b2 deficiency). Riboflavin exists as a solid, possibly soluble (in water), and an extremely weak basic (essentially neutral) compound (based on its pKa) molecule. Riboflavin exists in all living species, ranging from bacteria to humans. Riboflavin is a potentially toxic compound. |
|---|
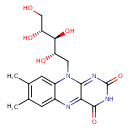
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)pentitol | ChEBI | | 6,7-Dimethyl-9-D-ribitylisoalloxazine | ChEBI | | 7,8-Dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)benzo[g]pteridine-2,4(3H,10H)-dione | ChEBI | | 7,8-Dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)isoalloxazine | ChEBI | | 7,8-Dimethyl-10-ribitylisoalloxazine | ChEBI | | e101 | ChEBI | | Lactoflavin | ChEBI | | Riboflavina | ChEBI | | RIBOFLAVINE | ChEBI | | Riboflavinum | ChEBI | | Vitamin b2 | ChEBI | | Vitamin g | ChEBI | | Bisulase | Kegg | | (-)-Riboflavin | HMDB | | 1-Deoxy-1-(3,4-dihydro-7,8-dimethyl-2,4-dioxobenzo[g]pteridin-10(2H)-yl)-D-ribitol | HMDB | | 6,7-Dimethyl-9-ribitylisoalloxazine | HMDB | | 7,8-Dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)-benzo[g]pteridine-2,4(3H,10H)-dione | HMDB | | Beflavin | HMDB | | Beflavine | HMDB | | Benzo[g]pteridine riboflavin deriv. | HMDB | | e 101 | HMDB | | Flavaxin | HMDB | | Flavin BB | HMDB | | Flaxain | HMDB | | FOOD Yellow 15 | HMDB | | Hyre | HMDB | | Lactobene | HMDB | | Lactoflavine | HMDB | | Ribipca | HMDB | | Ribocrisina | HMDB | | Riboderm | HMDB | | Ribosyn | HMDB | | Ribotone | HMDB | | Ribovel | HMDB | | Russupteridine yellow III | HMDB | | San yellow b | HMDB | | Vitaflavine | HMDB | | Vitasan b2 | HMDB | | Vitamin b 2 | HMDB |
|
|---|
| Chemical Formula | C17H20N4O6 |
|---|
| Average Molecular Weight | 376.3639 |
|---|
| Monoisotopic Molecular Weight | 376.138284392 |
|---|
| IUPAC Name | 7,8-dimethyl-10-[(2S,3S,4R)-2,3,4,5-tetrahydroxypentyl]-2H,3H,4H,10H-benzo[g]pteridine-2,4-dione |
|---|
| Traditional Name | riboflavin |
|---|
| CAS Registry Number | 83-88-5 |
|---|
| SMILES | CC1=C(C)C=C2N(C[C@H](O)[C@H](O)[C@H](O)CO)C3=NC(=O)NC(=O)C3=NC2=C1 |
|---|
| InChI Identifier | InChI=1S/C17H20N4O6/c1-7-3-9-10(4-8(7)2)21(5-11(23)14(25)12(24)6-22)15-13(18-9)16(26)20-17(27)19-15/h3-4,11-12,14,22-25H,5-6H2,1-2H3,(H,20,26,27)/t11-,12+,14-/m0/s1 |
|---|
| InChI Key | AUNGANRZJHBGPY-SCRDCRAPSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as flavins. Flavins are compounds containing a flavin (7,8-dimethyl-benzo[g]pteridine-2,4-dione) moiety, with a structure characterized by an isoalloaxzine tricyclic ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pteridines and derivatives |
|---|
| Sub Class | Alloxazines and isoalloxazines |
|---|
| Direct Parent | Flavins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Flavin
- Diazanaphthalene
- Quinoxaline
- Pyrimidone
- Pyrazine
- Pyrimidine
- Benzenoid
- Heteroaromatic compound
- Vinylogous amide
- Secondary alcohol
- Lactam
- Polyol
- Azacycle
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected and Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 290 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 0.0847 mg/mL | Not Available | | LogP | -1.46 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-006x-4980000000-dd278a577316361d270a | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-006x-4980000000-dd278a577316361d270a | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0btc-9014000000-75f046dc3c6cb008690e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-0zfs-5146149000-f9db57dd1ccd4a014604 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0a4i-0091000000-a82c54d3153103fcdb1f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0a4i-0091000000-a82c54d3153103fcdb1f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-0a4i-0090000000-2aff124ee1fc62c13844 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-0a4i-0090000000-f8b29c3e2c601a944a6c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-0a4i-0090000000-4e8c9bd38ea0f5ae9a94 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0090000000-5288e9226616bb75603c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0093000000-5999e10d04a53f4dae9b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0091000000-8f391045e25f26d53384 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-08fv-0290000000-b4d5f66b726c01d08c55 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0006-4690000000-d195fd5aee22ece7f2ec | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0039000000-549fbc0a59262f64680e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0195000000-58354696aaddaffc0fd8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0f76-9231000000-67715e21d6e9ce2655fe | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0a4i-0090000000-838afb0228d293bee0a3 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-2190000000-b659be001c9aa265c257 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0a4i-0090000000-21473bfb7f4a3eafd059 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-004i-0019000000-86365dedafa031aa7787 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0006-4390000000-ac1b59ab7cc2209f4241 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00dj-4900000000-72d33eb27b9bd6a13d9e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-004l-0569000000-874b71fdc78d04853bf0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-qTof , Positive | splash10-004i-0239000000-659ca9fae9643f3ce73d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-004l-0569000000-874b71fdc78d04853bf0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , positive | splash10-0006-0092000000-1f1be5508c1d50d8dff7 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , positive | splash10-0006-0092000000-74bf0b86efe72fe37198 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , positive | splash10-057i-0069000000-bb0522be4472e049dbc5 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| General References | - Agostini TS, Scherer R, Godoy HT: Simultaneous determination of B-group vitamins in enriched Brazilian dairy products. Crit Rev Food Sci Nutr. 2007;47(5):435-9. doi: 10.1080/10408390600846309. [PubMed:17558655 ]
- Gaucheron F: Milk and dairy products: a unique micronutrient combination. J Am Coll Nutr. 2011 Oct;30(5 Suppl 1):400S-9S. [PubMed:22081685 ]
- Park, Y. W; Juárez, Manuela ; Ramos, M.; Haenlein, G. F. W. (2007). Park, Y. W; Juárez, Manuela ; Ramos, M.; Haenlein, G. F. W.. Physico-chemical characteristics of goat and sheep milk. Small Ruminant Res.(2007) 68:88-113 doi: 10.1016/j.smallrumres.2006.09.013. Small Ruminant Research.
- A. Foroutan et al. (2019). A. Foroutan et al. The Chemical Composition of Commercial Cow's Milk (in preparation). Journal of Agricultural and Food Chemistry.
- USDA Food Composition Databases [Link]
- Fooddata+, The Technical University of Denmark (DTU) [Link]
|
|---|