| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:30:52 UTC |
|---|
| Update Date | 2020-04-21 18:14:07 UTC |
|---|
| BMDB ID | BMDB0000446 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | N-Alpha-ccetyllysine |
|---|
| Description | Acetyllysine belongs to the class of organic compounds known as n-acyl-alpha amino acids. N-acyl-alpha amino acids are compounds containing an alpha amino acid which bears an acyl group at its terminal nitrogen atom. Acetyllysine exists as a solid, possibly soluble (in water), and a very strong basic compound (based on its pKa) molecule. Acetyllysine exists in all eukaryotes, ranging from yeast to humans. |
|---|
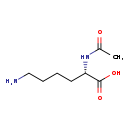
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2-(Acetylamino)-6-aminohexanoic acid | ChEBI | | N(alpha)-Acetyllysine | ChEBI | | N-Acetyl-L-lysine | ChEBI | | N2-Acetyl-L-lysine | ChEBI | | N(alpha)-Acetyl-L-lysine | ChEBI | | (2S)-2-(Acetylamino)-6-aminohexanoate | Generator | | N(a)-Acetyllysine | Generator | | N(Α)-acetyllysine | Generator | | N(a)-Acetyl-L-lysine | Generator | | N(Α)-acetyl-L-lysine | Generator | | N-a-Acetyl-L-lysine | Generator | | N-Α-acetyl-L-lysine | Generator | | 6-Amino-L-2-acetamidohexanoic acid | HMDB | | N2-Acetyllysine | HMDB | | Nα-acetyl-L-lysine | HMDB | | Nα-acetyllysine | HMDB | | Nalpha-acetyl-L-lysine | HMDB | | Nalpha-acetyllysine | HMDB | | N-a-Acetyllysine | HMDB | | N-Α-acetyllysine | HMDB | | 6-Amino-L-2-acetamidohexanoate | HMDB | | N-Acetyl poly-L-lysine | HMDB | | N-Acetyl polylysine | HMDB | | Acetyllysine | HMDB | | 6-Amino-2-[(1-hydroxyethylidene)amino]hexanoate | HMDB | | N(2)-Acetyllysine | HMDB | | N(alpha)-Acetyllysine, (DL)-isomer | HMDB | | N-alpha-Acetyllysine | HMDB | | N-alpha-Acetyl-L-lysine | ChEBI |
|
|---|
| Chemical Formula | C8H16N2O3 |
|---|
| Average Molecular Weight | 188.227 |
|---|
| Monoisotopic Molecular Weight | 188.116092383 |
|---|
| IUPAC Name | (2S)-6-amino-2-acetamidohexanoic acid |
|---|
| Traditional Name | nα-acetyllysine |
|---|
| CAS Registry Number | 1946-82-3 |
|---|
| SMILES | CC(=O)N[C@@H](CCCCN)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C8H16N2O3/c1-6(11)10-7(8(12)13)4-2-3-5-9/h7H,2-5,9H2,1H3,(H,10,11)(H,12,13)/t7-/m0/s1 |
|---|
| InChI Key | VEYYWZRYIYDQJM-ZETCQYMHSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as n-acyl-alpha amino acids. N-acyl-alpha amino acids are compounds containing an alpha amino acid which bears an acyl group at its terminal nitrogen atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | N-acyl-alpha amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-acyl-alpha-amino acid
- Medium-chain fatty acid
- Amino fatty acid
- Fatty acyl
- Fatty acid
- Amino acid
- Carboximidic acid
- Carboximidic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Amine
- Organooxygen compound
- Organonitrogen compound
- Primary amine
- Hydrocarbon derivative
- Primary aliphatic amine
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Carbonyl group
- Organic nitrogen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | Not Available |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-003i-1900000000-1bb977fd5835c87c7d01 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-9200000000-08d9b5954a220181ea5f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-001i-9000000000-9404d66c59d266164c23 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-052b-9600000000-64d02b65a9438fd24af1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0002-2900000000-4e4307890c5505c5aeba | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-3d476e678ca9d6df5ea4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-28b149c70053eaa89aa0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0002-0900000000-b154acfb571fb75b00f5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-003r-2900000000-b607df4d59ad27d4f5c3 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-003r-2900000000-bde2837d508c6909ec12 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000j-0900000000-0c503af7c1fbf54b2c2c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9700000000-cecd553985bf6c4d7160 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053r-9000000000-f705bda3174a35c518f8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-637ed07817e215db5233 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-1900000000-92bf1c9bbc3af7b68c0c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-57df45c6e237a09f2c4b | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 15.08 MHz, D2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|