| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:31:03 UTC |
|---|
| Update Date | 2020-05-11 20:00:38 UTC |
|---|
| BMDB ID | BMDB0000459 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
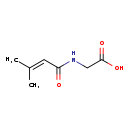
| Common Name | 3-Methylcrotonylglycine |
|---|
| Description | 3-Methylcrotonylglycine belongs to the class of organic compounds known as n-acyl-alpha amino acids. N-acyl-alpha amino acids are compounds containing an alpha amino acid which bears an acyl group at its terminal nitrogen atom. 3-Methylcrotonylglycine is an extremely weak basic (essentially neutral) compound (based on its pKa). 3-Methylcrotonylglycine is a potentially toxic compound. 3-Methylcrotonylglycine, with regard to humans, has been linked to several inborn metabolic disorders including biotinidase deficiency, 3-methyl-crotonyl-glycinuria, propionic acidemia, and 3-hydroxy-3-methylglutaryl-coa lyase deficiency. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| beta-Methylcrotonylglycine | ChEBI | | b-Methylcrotonylglycine | Generator | | Β-methylcrotonylglycine | Generator | | 3-Methylcrotonyl glycine | HMDB | | 3-Methylcrotonyl-glycine | HMDB | | 3-Methylcrotonylglycin | HMDB | | N-(3-Methyl-1-oxo-2-butenyl)-glycine | HMDB | | 3-Methylcrotonylglycine | MeSH |
|
|---|
| Chemical Formula | C7H11NO3 |
|---|
| Average Molecular Weight | 157.1671 |
|---|
| Monoisotopic Molecular Weight | 157.073893223 |
|---|
| IUPAC Name | 2-(3-methylbut-2-enamido)acetic acid |
|---|
| Traditional Name | (3-methylbut-2-enamido)acetic acid |
|---|
| CAS Registry Number | 33008-07-0 |
|---|
| SMILES | CC(C)=CC(=O)NCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C7H11NO3/c1-5(2)3-6(9)8-4-7(10)11/h3H,4H2,1-2H3,(H,8,9)(H,10,11) |
|---|
| InChI Key | PFWQSHXPNKRLIV-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as n-acyl-alpha amino acids. N-acyl-alpha amino acids are compounds containing an alpha amino acid which bears an acyl group at its terminal nitrogen atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | N-acyl-alpha amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-acyl-alpha-amino acid
- N-acyl-amine
- Secondary carboxylic acid amide
- Carboxamide group
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-001l-9200000000-b5de5c506d0864c0f7d1 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-01x3-9210000000-3cd3c3c8ea08c7d1e6c7 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-001i-9000000000-de6b64abad08a6b0aa93 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0a59-9000000000-6614867cbb98a3422626 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a59-9000000000-87fe2c98c5568e0a3651 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-053r-9000000000-6b621ffd0bb999f80e57 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-00di-9000000000-595a1236b9c6b885ba28 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-7900000000-f113c2d59fe1238ea573 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a6r-9100000000-ca7eb3d4b533d2e31892 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-78c1b06293f14b96b012 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-9462ca1f1e0bcf30b871 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-6900000000-7cf2a248f3b7490e7677 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ab9-9000000000-30da48e2c1ce30e69097 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-074i-5900000000-ec18735846b1c28e727c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9100000000-ca9e9c33cf4ebfc612fc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-9000000000-16027ff30f26d4efcd81 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03e9-9700000000-cec9fb8eb63cd434b7e8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a59-9000000000-33ae5682d1818bbe4123 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-93d70561c1a4ee06a679 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, CDCl3, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CD3OD, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Carter, S. M. Bonham; Watson, D. G.; Midgley, J. M.; Logan, R. W. Synthesis and characterization of acyl glycines. Their measurement in single blood spots by gas chromatography-mass spectrometry to diagnose inborn errors of metabolism. Journal of Chromatography, B: Biomedical Applications (1996), 677(1), 29-35. |
|---|