| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:32:13 UTC |
|---|
| Update Date | 2020-05-11 20:21:05 UTC |
|---|
| BMDB ID | BMDB0000527 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 6a,12a-Dihydroxylithocholic acid |
|---|
| Description | 6a,12a-Dihydroxylithocholic acid, also known as 3a,6a,12a-trihydroxy-5b-cholan-24-Oate, belongs to the class of organic compounds known as trihydroxy bile acids, alcohols and derivatives. These are prenol lipids structurally characterized by a bile acid or alcohol which bears three hydroxyl groups. Based on a literature review a significant number of articles have been published on 6a,12a-Dihydroxylithocholic acid. |
|---|
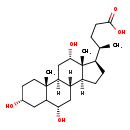
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 6a,12a-Dihydroxylithocholate | Generator | | 3a,6a,12a-Trihydroxy-5b-cholan-24-Oate | HMDB | | 3a,6a,12a-Trihydroxy-5b-cholan-24-Oic acid | HMDB | | 3a,6a,12a-Trihydroxycholate | HMDB | | 3a,6a,12a-Trihydroxycholic acid | HMDB | | (4R)-4-[(1S,2R,5R,8S,10R,11S,14R,15R,16S)-5,8,16-Trihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanoate | HMDB | | 6a,12a-Dihydroxylithocholic acid | Generator |
|
|---|
| Chemical Formula | C24H40O5 |
|---|
| Average Molecular Weight | 408.5714 |
|---|
| Monoisotopic Molecular Weight | 408.28757439 |
|---|
| IUPAC Name | (4R)-4-[(1S,2R,5R,8S,10R,11S,14R,15R,16S)-5,8,16-trihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanoic acid |
|---|
| Traditional Name | 3a,6a,12a-trihydroxycholate |
|---|
| CAS Registry Number | 21066-18-2 |
|---|
| SMILES | [H][C@@]12CC[C@H]([C@H](C)CCC(O)=O)[C@@]1(C)[C@@H](O)C[C@@]1([H])[C@@]2([H])C[C@H](O)C2C[C@H](O)CC[C@]12C |
|---|
| InChI Identifier | InChI=1S/C24H40O5/c1-13(4-7-22(28)29)16-5-6-17-15-11-20(26)19-10-14(25)8-9-23(19,2)18(15)12-21(27)24(16,17)3/h13-21,25-27H,4-12H2,1-3H3,(H,28,29)/t13-,14-,15+,16-,17+,18+,19?,20+,21+,23-,24-/m1/s1 |
|---|
| InChI Key | GCAHOAMXTYBLNZ-YAZNBHORSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as trihydroxy bile acids, alcohols and derivatives. These are prenol lipids structurally characterized by a bile acid or alcohol which bears three hydroxyl groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Trihydroxy bile acids, alcohols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Trihydroxy bile acid, alcohol, or derivatives
- 3-hydroxysteroid
- 12-hydroxysteroid
- 3-alpha-hydroxysteroid
- 6-hydroxysteroid
- Hydroxysteroid
- Cyclic alcohol
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Polyol
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Alcohol
- Organooxygen compound
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01ox-0329000000-4b46ad1b2dacce1bf218 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-001i-1100039000-760033c57e267918f963 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00dl-0009000000-f5b70cbc59d5ea066854 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dl-0009000000-a1598b92858405ff74e4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0931-2109000000-5040f3ce8f75f7259313 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4r-0009700000-8dc05bd9542a54f1f91b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-052r-0009200000-9a24af3f58b590ac86cd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9007000000-acd067cd1da85248dd66 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000900000-ec1d9319a9eaf1f2aba6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4r-0007900000-ee1a89f01c0c399bba19 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ly9-1019100000-1ab1ccc5b19c01372e0d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0abc-0009400000-c7e4fe766731000e676e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-059l-3249100000-f20fd02a8db7144f53cf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0aor-9630000000-8f0dedbb78fb6914fa90 | View in MoNA |
|---|
|
|---|
| Synthesis Reference | Iida, Takashi; Tamaru, Tamaaki; Chang, Frederic C.; Goto, Junichi; Nambara, Toshio. Potential bile acid metabolites. XVIII. Synthesis of stereoisomeric 3,6,12a-trihydroxy-5b-cholanoic acids. Journal of Lipid Research (1991), 32(4), 649-58. |
|---|