| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:33:24 UTC |
|---|
| Update Date | 2020-05-11 20:43:59 UTC |
|---|
| BMDB ID | BMDB0000600 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Galactosylhydroxylysine |
|---|
| Description | Galactosylhydroxylysine, also known as hydroxylysine-galactose, belongs to the class of organic compounds known as glycosyl-amino acids. Glycosyl-amino acids are compounds consisting of saccharide linked through a glycosyl linkage (O-, N-, or S-) to an amino acid. Based on a literature review a significant number of articles have been published on Galactosylhydroxylysine. |
|---|
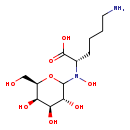
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 5-(beta-D-Galactopyranosyloxy)-L-lysine | HMDB | | 5-(beta-delta-Galactopyranosyloxy)-L-lysine | HMDB | | 5-O-beta-D-Galactopyranosylhydroxy-L-lysine | HMDB | | 5-O-beta-delta-Galactopyranosylhydroxy-L-lysine | HMDB | | beta-1-Galactosyl-O-hydroxylysine | HMDB | | Galactopyranosylhydroxy-L-lysine | HMDB | | Galactosyl-delta-hydroxylysine | HMDB | | Hydroxylysine-galactose | HMDB | | L-5-(beta-D-Galactopyranosyloxy)-lysine | HMDB | | L-5-(beta-delta-Galactopyranosyloxy)-lysine | HMDB | | O-beta-D-Galactopyranosylhydroxy-L-lysine | HMDB | | O-beta-delta-Galactopyranosylhydroxy-L-lysine | HMDB | | Procollagen (5-galactosyloxy)-L-lysine | HMDB | | (2S)-6-Amino-2-{n-hydroxy[(3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]amino}hexanoate | HMDB |
|
|---|
| Chemical Formula | C12H24N2O8 |
|---|
| Average Molecular Weight | 324.3276 |
|---|
| Monoisotopic Molecular Weight | 324.153265754 |
|---|
| IUPAC Name | (2S)-6-amino-2-{N-hydroxy[(3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]amino}hexanoic acid |
|---|
| Traditional Name | galactosylhydroxylysine |
|---|
| CAS Registry Number | 32448-36-5 |
|---|
| SMILES | NCCCC[C@H](N(O)C1O[C@H](CO)[C@H](O)[C@H](O)[C@H]1O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C12H24N2O8/c13-4-2-1-3-6(12(19)20)14(21)11-10(18)9(17)8(16)7(5-15)22-11/h6-11,15-18,21H,1-5,13H2,(H,19,20)/t6-,7+,8-,9-,10+,11?/m0/s1 |
|---|
| InChI Key | RCPOVANIIKXVTB-YPPRVYOWSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycosyl-amino acids. Glycosyl-amino acids are compounds consisting of saccharide linked through a glycosyl linkage (O-, N-, or S-) to an amino acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Glycosyl-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycosyl-amino-acid
- Hexose monosaccharide
- Glycosyl compound
- N-glycosyl compound
- N-hydroxyl-alpha-amino acid
- Alpha-amino acid or derivatives
- Medium-chain fatty acid
- Amino fatty acid
- Heterocyclic fatty acid
- Hydroxy fatty acid
- Monosaccharide
- Oxane
- Fatty acyl
- Fatty acid
- Secondary alcohol
- Amino acid
- Organoheterocyclic compound
- Polyol
- Carboxylic acid
- Oxacycle
- N-organohydroxylamine
- Monocarboxylic acid or derivatives
- Primary amine
- Primary aliphatic amine
- Alcohol
- Organonitrogen compound
- Hydrocarbon derivative
- Organooxygen compound
- Organic oxide
- Organopnictogen compound
- Carbonyl group
- Organic oxygen compound
- Primary alcohol
- Organic nitrogen compound
- Amine
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 257.0 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0pbl-9331000000-55f681ebb00dbb5b63ad | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (5 TMS) - 70eV, Positive | splash10-01c0-3922126000-4608c9d1673267935e9e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - CE-QTOF-MS system (Agilent 7100 CE + 6550 QTOF) 20V, Positive | splash10-03fs-0900000000-b277425c7b2baf2e1ce0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a6r-0319000000-e7265dc7d65b37c43370 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0c04-5934000000-12517f6dca52703813c3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dl-7910000000-7645fde658d953975315 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0692000000-ce838deeac1950cd5317 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0nu9-0961000000-0121d6fb23dab2a0d448 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0abc-6910000000-955acf234208503cf736 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0009000000-feb8033fade4ec36b163 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kb-1900000000-9887ff59cd8b2608215e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a59-9500000000-2e6b65bbf11b1b8a689f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0309000000-6517fbb0bf566b43f2d0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-5769000000-812154ea65e633cab3a0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00kf-4900000000-d6a34ed61253e7556f1d | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|