| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:35:51 UTC |
|---|
| Update Date | 2020-05-11 20:21:30 UTC |
|---|
| BMDB ID | BMDB0000743 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Hexacarboxylporphyrin I |
|---|
| Description | Hexacarboxylporphyrin I belongs to the class of organic compounds known as porphyrins. Porphyrins are compounds containing a fundamental skeleton of four pyrrole nuclei united through the alpha-positions by four methine groups to form a macrocyclic structure. Based on a literature review very few articles have been published on Hexacarboxylporphyrin I. |
|---|
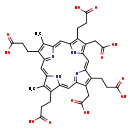
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Hexacarboxylic acid prophyrin I | HMDB | | Hexacarboxyporphyrin I | HMDB | | 3-[9,14,19-Tris(2-carboxyethyl)-10,15-bis(carboxymethyl)-5,20-dimethyl-21,22,23,24-tetraazapentacyclo[16.2.1.1³,⁶.1⁸,¹¹.1¹³,¹⁶]tetracosa-1,3(24),4,6,8,10,12,14,16(22),17,19-undecaen-4-yl]propanoate | HMDB |
|
|---|
| Chemical Formula | C38H38N4O12 |
|---|
| Average Molecular Weight | 742.7279 |
|---|
| Monoisotopic Molecular Weight | 742.2486227 |
|---|
| IUPAC Name | 3-[9,14,19-tris(2-carboxyethyl)-10,15-bis(carboxymethyl)-5,20-dimethyl-21,22,23,24-tetraazapentacyclo[16.2.1.1^{3,6}.1^{8,11}.1^{13,16}]tetracosa-1,3(24),4,6,8,10,12,14,16(22),17,19-undecaen-4-yl]propanoic acid |
|---|
| Traditional Name | 3-[9,14,19-tris(2-carboxyethyl)-10,15-bis(carboxymethyl)-5,20-dimethyl-21,22,23,24-tetraazapentacyclo[16.2.1.1^{3,6}.1^{8,11}.1^{13,16}]tetracosa-1,3(24),4,6,8,10,12,14,16(22),17,19-undecaen-4-yl]propanoic acid |
|---|
| CAS Registry Number | 73913-56-1 |
|---|
| SMILES | CC1=C(CCC(O)=O)/C2=C/C3=N/C(=C\C4=C(CC(O)=O)C(CCC(O)=O)=C(N4)/C=C4\N=C(\C=C\1/N\2)C(CCC(O)=O)=C4C)/C(CCC(O)=O)=C3CC(O)=O |
|---|
| InChI Identifier | InChI=1S/C38H38N4O12/c1-17-19(3-7-33(43)44)27-13-25-18(2)20(4-8-34(45)46)28(40-25)15-31-24(12-38(53)54)22(6-10-36(49)50)30(42-31)16-32-23(11-37(51)52)21(5-9-35(47)48)29(41-32)14-26(17)39-27/h13-16,40-41H,3-12H2,1-2H3,(H,43,44)(H,45,46)(H,47,48)(H,49,50)(H,51,52)(H,53,54)/b25-13-,26-14-,27-13-,28-15-,29-14-,30-16-,31-15-,32-16- |
|---|
| InChI Key | AFHLWIUSJBEMJE-ZMXCOFNRSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as porphyrins. Porphyrins are compounds containing a fundamental skeleton of four pyrrole nuclei united through the alpha-positions by four methine groups to form a macrocyclic structure. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Tetrapyrroles and derivatives |
|---|
| Sub Class | Porphyrins |
|---|
| Direct Parent | Porphyrins |
|---|
| Alternative Parents | Not Available |
|---|
| Substituents | Not Available |
|---|
| Molecular Framework | Not Available |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 208 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_16) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_17) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-056r-0000006900-c7b0f6210ebd3dd79245 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00or-0000009200-bf6d71def09551da3694 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0000009000-907019a41b59554e8fcc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00fv-0000008900-4d81ad3957c5de1f037c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00b9-0000009400-1129842b62b6eff09626 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a6r-4000009200-976122ebaaa0d6b1e75c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0000009000-946c7d847cb3aaba6995 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ufr-0000009000-9b6fee0babe743857350 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udi-0000009000-6af7dc488caae9f0b3e5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0000009600-40175966eda4e6eae1eb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f9i-0000009000-c841884267d2d4738e5e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udr-0000009000-d70a85e08a880ee035ad | View in MoNA |
|---|
|
|---|