| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:35:56 UTC |
|---|
| Update Date | 2020-04-22 15:05:07 UTC |
|---|
| BMDB ID | BMDB0000747 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Isovalerylalanine |
|---|
| Description | Isovalerylalanine, also known as isopentanoylalanine, belongs to the class of organic compounds known as n-acyl-l-alpha-amino acids. These are n-acylated alpha amino acids which have the L-configuration of the alpha-carbon atom. Based on a literature review a significant number of articles have been published on Isovalerylalanine. |
|---|
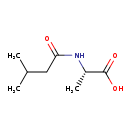
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2-(3-Methylbutanamido)propanoic acid | ChEBI | | Isopentanoylalanine | ChEBI | | N-Isopentanoyl-L-alanine | ChEBI | | N-Isopentanoylalanine | ChEBI | | N-Isovalerylalanine | ChEBI | | (2S)-2-(3-Methylbutanamido)propanoate | Generator | | N-Isovaleryl-L-alanine | HMDB | | Isovalerylalanine | ChEBI |
|
|---|
| Chemical Formula | C8H15NO3 |
|---|
| Average Molecular Weight | 173.2096 |
|---|
| Monoisotopic Molecular Weight | 173.105193351 |
|---|
| IUPAC Name | (2S)-2-(3-methylbutanamido)propanoic acid |
|---|
| Traditional Name | isovalerylalanine |
|---|
| CAS Registry Number | 68219-63-6 |
|---|
| SMILES | CC(C)CC(=O)N[C@@H](C)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C8H15NO3/c1-5(2)4-7(10)9-6(3)8(11)12/h5-6H,4H2,1-3H3,(H,9,10)(H,11,12)/t6-/m0/s1 |
|---|
| InChI Key | OJPSNARFDYTAEN-LURJTMIESA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as n-acyl-l-alpha-amino acids. These are n-acylated alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | N-acyl-L-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-acyl-l-alpha-amino acid
- Alanine or derivatives
- Fatty amide
- N-acyl-amine
- Fatty acyl
- Carboxamide group
- Secondary carboxylic acid amide
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Hydrocarbon derivative
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Organic oxygen compound
- Organic oxide
- Organopnictogen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000f-9200000000-0941d93d8b73095be371 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00bl-9310000000-ff31ec8bcd2f767edd93 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05i0-4900000000-279a1567e1a63ff40f7b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9200000000-75f940b78c284e853b5c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9000000000-84fea44c8d0eeaff9059 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-1900000000-c44e3552766c93b10335 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fki-7900000000-1dbfe9bbc6b7b04d91a6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05g3-9000000000-f37b9b82877a21760d9f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9300000000-bf002906104d17589e4f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9000000000-eee2c525ead39ccdd1b1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000f-9000000000-8f8a6b36163eb8d3cef2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-9200000000-ac4c7b8b294dbcda8355 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9000000000-49ba66e461e13d06ddf7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-7987039666b0de647c54 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 5%_DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Chen, Shu-Hui; Lamar, Jason; Guo, Deqi; Kohn, Todd; Yang, Hsiu-Chiung; McGee, James; Timm, David; Erickson, Jon; Yip, Yvonne; May, Patrick; McCarthy, James. P3 cap modified Phe*-Ala series BACE inhibitors. Bioorganic & Medicinal Chemistry Letters (2004), 14(1), 245-250. |
|---|