| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:37:32 UTC |
|---|
| Update Date | 2020-04-22 15:05:37 UTC |
|---|
| BMDB ID | BMDB0000853 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | N-Acetyl-b-D-galactosamine |
|---|
| Description | N-Acetyl-b-D-galactosamine, also known as beta-galnac or β-galnac, belongs to the class of organic compounds known as n-acyl-alpha-hexosamines. These are carbohydrate derivatives containing a hexose moiety in which the oxygen atom is replaced by an n-acyl group. N-Acetyl-b-D-galactosamine is an extremely weak basic (essentially neutral) compound (based on its pKa). N-Acetyl-b-D-galactosamine exists in all living organisms, ranging from bacteria to humans. |
|---|
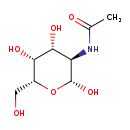
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Acetamido-2-deoxy-beta-D-galactopyranoside | ChEBI | | beta-GalNAc | ChEBI | | BGalNAc | ChEBI | | 2-Acetamido-2-deoxy-b-D-galactopyranoside | Generator | | 2-Acetamido-2-deoxy-β-D-galactopyranoside | Generator | | b-GalNAc | Generator | | Β-galnac | Generator | | 2-Deoxy-2-acetamido-b-D-galactopyranose | HMDB | | 2-Deoxy-2-acetamido-beta-D-galactopyranose | HMDB | | 2-Deoxy-2-acetamido-beta-delta-galactopyranose | HMDB | | b-D-2-Acetamido-2-deoxy-galactopyranose | HMDB | | b-N-Acetyl-D-galactosamine | HMDB | | b-N-Acetylgalactosamine | HMDB | | beta-D-2-Acetamido-2-deoxy-galactopyranose | HMDB | | beta-delta-2-Acetamido-2-deoxy-galactopyranose | HMDB | | beta-N-Acetyl-D-galactosamine | HMDB | | beta-N-Acetyl-delta-galactosamine | HMDB | | beta-N-Acetylgalactosamine | HMDB | | 2 Acetamido 2 D galactopyranose | HMDB | | 2-Acetamido-2-deoxy-D-galactose | HMDB | | 2 Acetamido 2 deoxy D galactose | HMDB | | 2-Acetamido-2-D-galactopyranose | HMDB | | Acetylgalactosamine | HMDB | | 2 Acetamido 2 deoxygalactose | HMDB | | N-Acetyl-D-galactosamine | HMDB | | 2-Acetamido-2-deoxygalactose | HMDB | | N Acetyl D galactosamine | HMDB | | N-Acetyl-β-D-galactosamine | HMDB | | N-Acetyl-b-D-galactosamine | Generator |
|
|---|
| Chemical Formula | C8H15NO6 |
|---|
| Average Molecular Weight | 221.2078 |
|---|
| Monoisotopic Molecular Weight | 221.089937217 |
|---|
| IUPAC Name | N-[(2R,3R,4R,5R,6R)-2,4,5-trihydroxy-6-(hydroxymethyl)oxan-3-yl]acetamide |
|---|
| Traditional Name | N-acetyl-β-D-galactosamine |
|---|
| CAS Registry Number | 14131-60-3 |
|---|
| SMILES | CC(=O)N[C@H]1[C@H](O)O[C@H](CO)[C@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C8H15NO6/c1-3(11)9-5-7(13)6(12)4(2-10)15-8(5)14/h4-8,10,12-14H,2H2,1H3,(H,9,11)/t4-,5-,6+,7-,8-/m1/s1 |
|---|
| InChI Key | OVRNDRQMDRJTHS-JAJWTYFOSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as n-acyl-alpha-hexosamines. These are carbohydrate derivatives containing a hexose moiety in which the oxygen atom is replaced by an n-acyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | N-acyl-alpha-hexosamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-acyl-alpha-hexosamine
- Hexose monosaccharide
- Monosaccharide
- Oxane
- Hemiacetal
- Secondary alcohol
- Carboximidic acid
- Carboximidic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Alcohol
- Organonitrogen compound
- Organic nitrogen compound
- Primary alcohol
- Organopnictogen compound
- Hydrocarbon derivative
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ue9-6920000000-c07df80c6fb758af9cd2 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-0007-3331900000-c6fab10d67507eecce65 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-1490000000-40dfe33adabe6ad7a7a8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0il0-2940000000-880c7b1fb58ed63f0ee2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01ox-9400000000-e1f07558d7921e9f24e1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-05g0-8920000000-d504941e0508d1593f33 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0pb9-9820000000-753556be259b7e745ba7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9100000000-bad102d081eb3bbc006b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0390000000-3820a7eec5a8c72f49d9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0h90-9740000000-db880363194a3979c637 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0229-9200000000-cdcf015aa799dc7d6d8b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-9500000000-2c2b09cb6bfb29039085 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0abc-9540000000-a1f7d557237a521d306a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-18eabe9e3d3657888360 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Chaplin, David; Crout, David H. G.; Bornemann, Stephen; Hutchinson, David W.; Khan, Riaz. Conversion of 2-acetamido-2-deoxy-b-D-glucopyranose (N-acetylglucosamine) into 2-acetamido-2-deoxy-b-D-galactopyranose (N-acetylgalactosamine using a biotransformation to generate a selectively deprotected substrate for SN2 inversion. Journal of the Chemical Society, Perkin Transactions 1: Organic and Bio-Organic Chemistry (1972-1999) (1992), (2), 235-7. |
|---|