| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:39:46 UTC |
|---|
| Update Date | 2020-04-22 15:06:17 UTC |
|---|
| BMDB ID | BMDB0001005 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
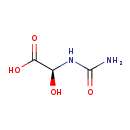
| Common Name | (S)-Ureidoglycolic acid |
|---|
| Description | (S)-Ureidoglycolic acid, also known as (S)-ureidoglycolate, belongs to the class of organic compounds known as n-carbamoyl-alpha amino acids. N-carbamoyl-alpha amino acids are compounds containing an alpha amino acid which bears an carbamoyl group at its terminal nitrogen atom (S)-Ureidoglycolic acid exists in all living species, ranging from bacteria to plants to humans. Based on a literature review very few articles have been published on (S)-Ureidoglycolic acid. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (-)-Ureidoglycolate | ChEBI | | (S)-Ureidoglycolate | ChEBI | | (-)-Ureidoglycolic acid | Generator | | (S)-[(Aminocarbonyl)amino]hydroxy-acetic acid | HMDB | | S-(-)-Ureidoglycolic acid | HMDB | | Ureidoglycolate | HMDB |
|
|---|
| Chemical Formula | C3H6N2O4 |
|---|
| Average Molecular Weight | 134.0907 |
|---|
| Monoisotopic Molecular Weight | 134.03275669 |
|---|
| IUPAC Name | (2S)-2-(carbamoylamino)-2-hydroxyacetic acid |
|---|
| Traditional Name | ureidoglycolate |
|---|
| CAS Registry Number | 7424-03-5 |
|---|
| SMILES | NC(=O)N[C@@H](O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C3H6N2O4/c4-3(9)5-1(6)2(7)8/h1,6H,(H,7,8)(H3,4,5,9)/t1-/m0/s1 |
|---|
| InChI Key | NWZYYCVIOKVTII-SFOWXEAESA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as n-carbamoyl-alpha amino acids. N-carbamoyl-alpha amino acids are compounds containing an alpha amino acid which bears an carbamoyl group at its terminal nitrogen atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | N-carbamoyl-alpha amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-carbamoyl-alpha-amino acid
- Alpha-hydroxy acid
- Hydroxy acid
- Carbonic acid derivative
- Urea
- Alkanolamine
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxide
- Organopnictogen compound
- Organic nitrogen compound
- Organic oxygen compound
- Carbonyl group
- Organonitrogen compound
- Organooxygen compound
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01r6-9100000000-a7e6b112e52795850588 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-02mi-8920000000-76af510166ac80995acd | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-9700000000-3a4e2e6ced0b05835ca9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dl-9000000000-f71cc7c577d24a593825 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01ox-9000000000-8e08566417cf9479b434 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9100000000-b163bec3ba10c56f6646 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-052f-9000000000-9777b17cbceaafa658b0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-e546d2761ff46042436b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9100000000-f83fb023b339c1a74e1a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-10d946d055df2a11aace | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-90726b17dc36e29c5299 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-9500000000-098f8ca862d2f98d80ad | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0h2f-9200000000-2b972ed857e447394a61 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-940f7cfcfa787f6857ae | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|