| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:39:53 UTC |
|---|
| Update Date | 2020-05-11 20:21:59 UTC |
|---|
| BMDB ID | BMDB0001013 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Cotinineglucuronide |
|---|
| Description | Cotinine glucuronide belongs to the class of organic compounds known as n-glucuronides. These are glucuronides in which the aglycone is linked to the carbohydrate unit through a N-glycosidic bond. Based on a literature review a significant number of articles have been published on Cotinine glucuronide. |
|---|
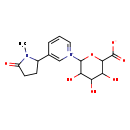
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-b-D-Glucopyranuronosyl-3-(1-methyl-5-oxo-2-pyrrolidinyl)- pyridinium | HMDB | | 1-beta-delta-Glucopyranuronosyl-3-(1-methyl-5-oxo-2-pyrrolidinyl)- pyridinium | HMDB | | Cotinine-glucuronide | HMDB | | Cotinine-N-glucuronide | HMDB |
|
|---|
| Chemical Formula | C16H20N2O7 |
|---|
| Average Molecular Weight | 352.3392 |
|---|
| Monoisotopic Molecular Weight | 352.127051004 |
|---|
| IUPAC Name | 1-(6-carboxylato-3,4,5-trihydroxyoxan-2-yl)-3-(1-methyl-5-oxopyrrolidin-2-yl)-1λ⁵-pyridin-1-ylium |
|---|
| Traditional Name | 1-(6-carboxylato-3,4,5-trihydroxyoxan-2-yl)-3-(1-methyl-5-oxopyrrolidin-2-yl)-1λ⁵-pyridin-1-ylium |
|---|
| CAS Registry Number | 139427-57-9 |
|---|
| SMILES | CN1C(CCC1=O)C1=C[N+](=CC=C1)C1OC(C(O)C(O)C1O)C([O-])=O |
|---|
| InChI Identifier | InChI=1S/C16H20N2O7/c1-17-9(4-5-10(17)19)8-3-2-6-18(7-8)15-13(22)11(20)12(21)14(25-15)16(23)24/h2-3,6-7,9,11-15,20-22H,4-5H2,1H3 |
|---|
| InChI Key | XWZCZWKUGIQPJD-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as n-glucuronides. These are glucuronides in which the aglycone is linked to the carbohydrate unit through a N-glycosidic bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | N-glucuronides |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-glucuronide
- Glycosyl compound
- N-glycosyl compound
- Pyrrolidinylpyridine
- Alkaloid or derivatives
- Beta-hydroxy acid
- Hydroxy acid
- Oxane
- Pyran
- Pyridine
- Pyridinium
- Pyrrolidone
- 2-pyrrolidone
- N-alkylpyrrolidine
- Heteroaromatic compound
- Tertiary carboxylic acid amide
- Pyrrolidine
- Carboxylic acid salt
- Carboxamide group
- Secondary alcohol
- Lactam
- Azacycle
- Oxacycle
- Organoheterocyclic compound
- Carboxylic acid
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Polyol
- Organic oxide
- Organopnictogen compound
- Carbonyl group
- Alcohol
- Organic nitrogen compound
- Organic salt
- Hydrocarbon derivative
- Organonitrogen compound
- Organic zwitterion
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | Not Available |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-9164000000-b507b999fbd5fca0f223 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-000f-6495530000-21635971da54dc89194c | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_3_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0009000000-066161aa1f36ad6f663f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f6t-3109000000-c3e5406589413be5cb13 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004i-8749000000-e6aa316b9fd267e9d5a1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0009000000-f8f14b208ec561f6a6ff | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0zfr-7649000000-5427908cfab7b2870e1a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9200000000-dc04c7ca765a4e1d44ae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0409000000-d21a6b67413145b1097e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-0925000000-700f183e58bc9857e5eb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004i-2920000000-d669575d2e816336cf4d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0119000000-dab9c1825610098e6840 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-2798000000-40dc27d7112d03e4133c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0072-2944000000-35c7f2cec72fddf72f5c | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|