| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:40:41 UTC |
|---|
| Update Date | 2020-05-11 20:38:36 UTC |
|---|
| BMDB ID | BMDB0001067 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | N-Acetylaspartylglutamic acid |
|---|
| Description | N-Acetylaspartylglutamic acid, also known as a-spaglumic acid or NAAG, belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. N-Acetylaspartylglutamic acid is an extremely weak basic (essentially neutral) compound (based on its pKa). |
|---|
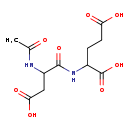
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| N-Acetylaspartylglutamate | Generator | | 2-({3-carboxy-1-hydroxy-2-[(1-hydroxyethylidene)amino]propylidene}amino)pentanedioate | HMDB | | a-Spaglumic acid | HMDB | | Acetyl-a-L-aspartylglutamic acid | HMDB | | Acetyl-alpha-L-aspartylglutamic acid | HMDB | | alpha-Spaglumic acid | HMDB | | Isospaglumic acid | HMDB | | N-(N-Acetylaspartyl)glutamic acid | HMDB | | N-Acetyl-a-aspartylglutamic acid | HMDB | | N-Acetyl-a-L-aspartyl-L-glutamic acid | HMDB | | N-Acetyl-alpha-aspartylglutamic acid | HMDB | | N-Acetyl-alpha-L-aspartyl-L-glutamic acid | HMDB | | N-Acetyl-L-aspartyl-L-glutamic acid | HMDB | | NAAG | HMDB |
|
|---|
| Chemical Formula | C11H16N2O8 |
|---|
| Average Molecular Weight | 304.2533 |
|---|
| Monoisotopic Molecular Weight | 304.090665498 |
|---|
| IUPAC Name | 2-(3-carboxy-2-acetamidopropanamido)pentanedioic acid |
|---|
| Traditional Name | 2-(3-carboxy-2-acetamidopropanamido)pentanedioic acid |
|---|
| CAS Registry Number | 3106-85-2 |
|---|
| SMILES | CC(=O)NC(CC(O)=O)C(=O)NC(CCC(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C11H16N2O8/c1-5(14)12-7(4-9(17)18)10(19)13-6(11(20)21)2-3-8(15)16/h6-7H,2-4H2,1H3,(H,12,14)(H,13,19)(H,15,16)(H,17,18)(H,20,21) |
|---|
| InChI Key | OPVPGKGADVGKTG-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Dipeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-dipeptide
- Glutamic acid or derivatives
- Aspartic acid or derivatives
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- Alpha-amino acid amide
- N-substituted-alpha-amino acid
- Alpha-amino acid or derivatives
- Tricarboxylic acid or derivatives
- Fatty amide
- Fatty acyl
- N-acyl-amine
- Acetamide
- Carboxamide group
- Secondary carboxylic acid amide
- Carboxylic acid
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Organonitrogen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0540-9880000000-c7c9c0da9dbe98eab286 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-05fr-9461560000-ce0ada373d3e90e6c80c | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0002-0900000000-2f2be5e8f2f3cf8bb047 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001r-9600000000-b3b345b7a22f2cdfac08 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-001r-9000000000-20ed6fe2510cf8b50b0a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0190000000-69dcaac6a427428064b4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f8j-3890000000-205f74a5dd8b3075e884 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0019-9400000000-743ec6872c52fd64b45d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0zg3-0193000000-467da8775363ff392ab6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05mx-1490000000-0aeed33e0dd1146b31de | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9710000000-4fd638435c8a9ed8fc4b | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, CD3OD, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CD3OD, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Marchetti, Enzo; Mattalia, Gabriele; Curatolo, Francesco; Bergesi, Giuseppe. Structure and synthesis of the natural dipeptides N-acetyl-b-L-aspartyl-L-glutamic acid and its isomer N-acetyl-a-L-aspartyl-L-glutamic acid. Annali di Chimica (Rome, Italy) (1967), 57(6), 624-31. |
|---|