| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:40:46 UTC |
|---|
| Update Date | 2020-05-21 16:28:33 UTC |
|---|
| BMDB ID | BMDB0001076 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Fructose 1-phosphate |
|---|
| Description | Fructose 1-phosphate, also known as D-fructose-1-P, belongs to the class of organic compounds known as hexose phosphates. These are carbohydrate derivatives containing a hexose substituted by one or more phosphate groups. Fructose 1-phosphate is possibly soluble (in water) and an extremely weak basic (essentially neutral) compound (based on its pKa). Fructose 1-phosphate exists in all living organisms, ranging from bacteria to humans. Fructose 1-phosphate participates in a number of enzymatic reactions, within cattle. In particular, Fructose 1-phosphate can be biosynthesized from D-fructose through the action of the enzyme ketohexokinase. In addition, Fructose 1-phosphate can be converted into dihydroxyacetone phosphate and glyceraldehyde; which is catalyzed by the enzyme fructose-bisphosphate aldolase a. In cattle, fructose 1-phosphate is involved in the metabolic pathway called the fructose and mannose degradation pathway. |
|---|
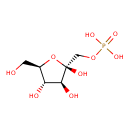
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| beta-D-Fructose 1-phosphate | ChEBI | | b-D-Fructose 1-phosphate | Generator | | b-D-Fructose 1-phosphoric acid | Generator | | beta-D-Fructose 1-phosphoric acid | Generator | | Β-D-fructose 1-phosphate | Generator | | Β-D-fructose 1-phosphoric acid | Generator | | Fructose 1-phosphoric acid | Generator | | D-Fructose 1-phosphate | HMDB | | D-Fructose-1-p | HMDB | | D-Fructose-1-phosphate | HMDB | | Fructose-1-p | HMDB | | Fructose-1-phosphate | HMDB | | b-D-Fructofuranose 1-phosphate | Generator | | b-D-Fructofuranose 1-phosphoric acid | Generator | | beta-D-Fructofuranose 1-phosphoric acid | Generator | | Β-D-fructofuranose 1-phosphate | Generator | | Β-D-fructofuranose 1-phosphoric acid | Generator |
|
|---|
| Chemical Formula | C6H13O9P |
|---|
| Average Molecular Weight | 260.1358 |
|---|
| Monoisotopic Molecular Weight | 260.029718526 |
|---|
| IUPAC Name | {[(2R,3S,4S,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)oxolan-2-yl]methoxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3S,4S,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)oxolan-2-yl]methoxyphosphonic acid |
|---|
| CAS Registry Number | 15978-08-2 |
|---|
| SMILES | OC[C@H]1O[C@](O)(COP(O)(O)=O)[C@@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C6H13O9P/c7-1-3-4(8)5(9)6(10,15-3)2-14-16(11,12)13/h3-5,7-10H,1-2H2,(H2,11,12,13)/t3-,4-,5+,6-/m1/s1 |
|---|
| InChI Key | RHKKZBWRNHGJEZ-ARQDHWQXSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hexose phosphates. These are carbohydrate derivatives containing a hexose substituted by one or more phosphate groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Hexose phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hexose phosphate
- Pentose phosphate
- Pentose-5-phosphate
- C-glycosyl compound
- Glycosyl compound
- Monosaccharide phosphate
- Pentose monosaccharide
- Monoalkyl phosphate
- Organic phosphoric acid derivative
- Alkyl phosphate
- Phosphoric acid ester
- Tetrahydrofuran
- Secondary alcohol
- Hemiacetal
- Organoheterocyclic compound
- Oxacycle
- Polyol
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Primary alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03di-9510000000-71d828159b61f8342118 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-001i-2912320000-08ffe3d23f5fa5d6162c | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dm-9470000000-18ac582a370338a3f5f1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01oy-7390000000-1a5a72af8bb734e50eb8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-009y-9100000000-4f151f373d7730140f38 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a6r-9270000000-5af91634007091008210 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-5cf3f457b8c6f6270b68 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-4e6feccf18943de8c419 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-1090000000-03030e5df4374fea160c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-9140000000-be637d8fd4ff371117c8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9010000000-26f483248da003aafbdb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0190000000-6beaa046be46044471be | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01p2-6920000000-7fd7dcf4a4f1479d91b1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052b-9100000000-60f4d9de70c1b7a7e1de | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| General References | - Melzer N, Wittenburg D, Hartwig S, Jakubowski S, Kesting U, Willmitzer L, Lisec J, Reinsch N, Repsilber D: Investigating associations between milk metabolite profiles and milk traits of Holstein cows. J Dairy Sci. 2013 Mar;96(3):1521-34. doi: 10.3168/jds.2012-5743. [PubMed:23438684 ]
|
|---|