| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:44:23 UTC |
|---|
| Update Date | 2020-05-21 16:28:52 UTC |
|---|
| BMDB ID | BMDB0001340 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Diguanosine tetraphosphate |
|---|
| Description | Diguanosine tetraphosphate, also known as GPPPPG or (PPG)2, belongs to the class of organic compounds known as (5'->5')-dinucleotides. These are dinucleotides where the two bases are connected via a (5'->5')-phosphodiester linkage. Diguanosine tetraphosphate is a moderately basic compound (based on its pKa). Diguanosine tetraphosphate exists in all living species, ranging from bacteria to humans. |
|---|
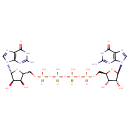
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (PPG)2 | ChEBI | | Bis(5'-guanosyl) tetraphosphate | ChEBI | | Bis(guanylyl) diphosphate | ChEBI | | g(5')P4(5')g | ChEBI | | GP4g | ChEBI | | GPPPPG | ChEBI | | Guanosine(5')tetraphospho(5')guanosine | ChEBI | | P1,P4-Bis(5'-guanosyl) tetraphosphate | ChEBI | | Bis(5'-guanosyl) tetraphosphoric acid | Generator | | Bis(guanylyl) diphosphoric acid | Generator | | P1,P4-Bis(5'-guanosyl) tetraphosphoric acid | Generator | | Diguanosine tetraphosphoric acid | Generator | | [5-(2-Amino-6-hydroxy-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-[[[[5-(2-amino-6-hydroxy-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-phosphinic acid | HMDB |
|
|---|
| Chemical Formula | C20H28N10O21P4 |
|---|
| Average Molecular Weight | 868.3858 |
|---|
| Monoisotopic Molecular Weight | 868.038094056 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(2-amino-6-oxo-6,9-dihydro-1H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}({[({[({[(2R,3S,4R,5R)-5-(2-amino-6-oxo-6,9-dihydro-1H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)(hydroxy)phosphoryl]oxy})phosphinic acid |
|---|
| Traditional Name | diguanosine tetraphosphate |
|---|
| CAS Registry Number | 4130-19-2 |
|---|
| SMILES | NC1=NC2=C(N=CN2[C@@H]2O[C@H](COP(O)(=O)OP(O)(=O)OP(O)(=O)OP(O)(=O)OC[C@H]3O[C@H]([C@H](O)[C@@H]3O)N3C=NC4=C3N=C(N)NC4=O)[C@@H](O)[C@H]2O)C(=O)N1 |
|---|
| InChI Identifier | InChI=1S/C20H28N10O21P4/c21-19-25-13-7(15(35)27-19)23-3-29(13)17-11(33)9(31)5(47-17)1-45-52(37,38)49-54(41,42)51-55(43,44)50-53(39,40)46-2-6-10(32)12(34)18(48-6)30-4-24-8-14(30)26-20(22)28-16(8)36/h3-6,9-12,17-18,31-34H,1-2H2,(H,37,38)(H,39,40)(H,41,42)(H,43,44)(H3,21,25,27,35)(H3,22,26,28,36)/t5-,6-,9-,10-,11-,12-,17-,18-/m1/s1 |
|---|
| InChI Key | OLGWXCQXRSSQPO-MHARETSRSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as (5'->5')-dinucleotides. These are dinucleotides where the two bases are connected via a (5'->5')-phosphodiester linkage. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | (5'->5')-dinucleotides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | (5'->5')-dinucleotides |
|---|
| Alternative Parents | |
|---|
| Substituents | - (5'->5')-dinucleotide
- Purine ribonucleoside polyphosphate
- Purine nucleotide sugar
- Purine ribonucleoside monophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-oxopurine
- Hypoxanthine
- Monosaccharide phosphate
- Imidazopyrimidine
- Purine
- Pyrimidone
- Monoalkyl phosphate
- Aminopyrimidine
- Pyrimidine
- Alkyl phosphate
- Phosphoric acid ester
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Tetrahydrofuran
- Vinylogous amide
- Azole
- Imidazole
- Heteroaromatic compound
- Secondary alcohol
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Organic nitrogen compound
- Amine
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Primary amine
- Alcohol
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0910110030-a6bd659122b84da65d3f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-c04b5e08bc949915c2fd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-0900000000-de8f07c2ed0e9f17f0ee | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0gb9-0700011090-f69c78a4b50e25f0eb5c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0900010000-b766f54c386c57ea4bcd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udi-2915010000-506add7a25f1233ab648 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000020-b1759ae61121c874c253 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000020-ad098ec0ac59f44f6bc6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udr-0900100000-91fc747e55e03728ef79 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000000090-cb93d23c88ced7dae5b0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0gb9-0400020390-175e627e05080096994a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pbc-0510390050-039fd4c9138a3432cc17 | View in MoNA |
|---|
| MS | Mass Spectrum (Electron Ionization) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|