| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:44:29 UTC |
|---|
| Update Date | 2020-05-21 16:28:32 UTC |
|---|
| BMDB ID | BMDB0001346 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | GDP-4-Dehydro-6-deoxy-D-mannose |
|---|
| Description | GDP-4-Dehydro-6-deoxy-D-mannose, also known as GDP-4-dehydro-D-rhamnose or GDP-4-keto-6-deoxymannose, belongs to the class of organic compounds known as purine nucleotide sugars. These are purine nucleotides bound to a saccharide derivative through the terminal phosphate group. GDP-4-Dehydro-6-deoxy-D-mannose is possibly soluble (in water) and a strong basic compound (based on its pKa). GDP-4-Dehydro-6-deoxy-D-mannose exists in all living organisms, ranging from bacteria to humans. GDP-4-Dehydro-6-deoxy-D-mannose participates in a number of enzymatic reactions, within cattle. In particular, GDP-4-Dehydro-6-deoxy-D-mannose can be biosynthesized from guanosine diphosphate mannose through its interaction with the enzyme GDP-mannose 4,6 dehydratase. In addition, GDP-4-Dehydro-6-deoxy-D-mannose can be biosynthesized from GDP-L-fucose; which is mediated by the enzyme GDP-L-fucose synthase. In cattle, GDP-4-dehydro-6-deoxy-D-mannose is involved in the metabolic pathway called the fructose and mannose degradation pathway. |
|---|
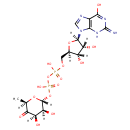
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| GDP-4-Dehydro-6-deoxy-D-talose | ChEBI | | GDP-4-Dehydro-D-rhamnose | ChEBI | | GDP-4-Keto-6-deoxy-D-mannose | ChEBI | | GDP-4-Keto-6-deoxymannose | ChEBI | | GDP-4-oxo-6-Deoxy-D-mannose | ChEBI | | Guanosine 5'-(trihydrogen diphosphate), p'-(6-deoxy-alpha-D-lyxo-hexopyranos-4-ulos-1-yl) ester | ChEBI | | Guanosine diphosphate-4-keto-6-deoxy-D-mannose | ChEBI | | GDP-4-Dehydro-alpha-D-rhamnose | Kegg | | GDP-4-Dehydro-6-deoxy-alpha-D-mannose | Kegg | | Guanosine 5'-(trihydrogen diphosphate), p'-(6-deoxy-a-D-lyxo-hexopyranos-4-ulos-1-yl) ester | Generator | | Guanosine 5'-(trihydrogen diphosphate), p'-(6-deoxy-α-D-lyxo-hexopyranos-4-ulos-1-yl) ester | Generator | | Guanosine 5'-(trihydrogen diphosphoric acid), p'-(6-deoxy-a-D-lyxo-hexopyranos-4-ulos-1-yl) ester | Generator | | Guanosine 5'-(trihydrogen diphosphoric acid), p'-(6-deoxy-alpha-D-lyxo-hexopyranos-4-ulos-1-yl) ester | Generator | | Guanosine 5'-(trihydrogen diphosphoric acid), p'-(6-deoxy-α-D-lyxo-hexopyranos-4-ulos-1-yl) ester | Generator | | Guanosine diphosphoric acid-4-keto-6-deoxy-D-mannose | Generator | | GDP-4-Dehydro-a-D-rhamnose | Generator | | GDP-4-Dehydro-α-D-rhamnose | Generator | | GDP-4-Dehydro-6-deoxy-a-D-mannose | Generator | | GDP-4-Dehydro-6-deoxy-α-D-mannose | Generator | | GDP-4-Keto-6-D-deoxymannose | HMDB | | GDP-4-oxo-6-Deoxymannose | HMDB | | Guanosine 5'-diphosphate 4-keto-6-deoxy-D-mannose | HMDB | | Guanosine 5’-diphosphate 4-keto-6-deoxy-D-mannose | HMDB |
|
|---|
| Chemical Formula | C16H23N5O15P2 |
|---|
| Average Molecular Weight | 587.3258 |
|---|
| Monoisotopic Molecular Weight | 587.066588115 |
|---|
| IUPAC Name | [({[(2R,3S,4R,5R)-5-(2-amino-6-oxo-6,9-dihydro-1H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2R,3S,4R,6R)-3,4-dihydroxy-6-methyl-5-oxooxan-2-yl]oxy})phosphinic acid |
|---|
| Traditional Name | gdp-4-keto-6-deoxymannose |
|---|
| CAS Registry Number | 18186-48-6 |
|---|
| SMILES | C[C@H]1O[C@H](OP(O)(=O)OP(O)(=O)OC[C@H]2O[C@H]([C@H](O)[C@@H]2O)N2C=NC3=C2N=C(N)NC3=O)[C@@H](O)[C@@H](O)C1=O |
|---|
| InChI Identifier | InChI=1S/C16H23N5O15P2/c1-4-7(22)9(24)11(26)15(33-4)35-38(30,31)36-37(28,29)32-2-5-8(23)10(25)14(34-5)21-3-18-6-12(21)19-16(17)20-13(6)27/h3-5,8-11,14-15,23-26H,2H2,1H3,(H,28,29)(H,30,31)(H3,17,19,20,27)/t4-,5-,8-,9+,10-,11+,14-,15-/m1/s1 |
|---|
| InChI Key | PNHLMHWWFOPQLK-BKUUWRAGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine nucleotide sugars. These are purine nucleotides bound to a saccharide derivative through the terminal phosphate group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine nucleotide sugars |
|---|
| Direct Parent | Purine nucleotide sugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine nucleotide sugar
- Purine ribonucleoside diphosphate
- Purine ribonucleoside monophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-oxopurine
- Hypoxanthine
- Monosaccharide phosphate
- Organic pyrophosphate
- Imidazopyrimidine
- Purine
- Aminopyrimidine
- Monoalkyl phosphate
- Pyrimidone
- Pyrimidine
- Alkyl phosphate
- Phosphoric acid ester
- Oxane
- Monosaccharide
- Organic phosphoric acid derivative
- N-substituted imidazole
- Tetrahydrofuran
- Vinylogous amide
- Azole
- Imidazole
- Heteroaromatic compound
- Cyclic ketone
- Secondary alcohol
- Ketone
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Hydrocarbon derivative
- Primary amine
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Carbonyl group
- Alcohol
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00mo-7594640000-f6eaffb3b87a7209c04d | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-002f-6891604000-2d6996a185dcede1dcba | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("GDP-4-Dehydro-6-deoxy-D-mannose,1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0910520000-fcbc13f148251be94ff4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0910000000-ed550a57c125fee026ee | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-1900000000-a873b451fb06d591cb14 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0f79-3901480000-ab8899450fb110bccaf6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-1901000000-c6a3e1a0d5247f947a8a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pdl-3900000000-148e1fc1e5c7ae57395c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000090000-45b44004cbf29fe6aa8f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-072c-7902450000-9b1901584b05fbc14f34 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05fr-2721910000-5a19390a8eaa546bc477 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900030000-a9251fc5a1b28bbacc54 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-2911220000-4e86fd74cab914f8c823 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0zgr-9631000000-4a357d49912c73e87853 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|