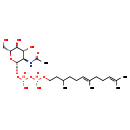| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:46:06 UTC |
|---|
| Update Date | 2020-04-22 15:08:14 UTC |
|---|
| BMDB ID | BMDB0001445 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | N-Acetyl-D-glucosaminyldiphosphodolichol |
|---|
| Description | N-Acetyl-D-glucosaminyldiphosphodolichol belongs to the class of organic compounds known as n-acyl-alpha-hexosamines. These are carbohydrate derivatives containing a hexose moiety in which the oxygen atom is replaced by an n-acyl group. N-Acetyl-D-glucosaminyldiphosphodolichol is an extremely weak basic (essentially neutral) compound (based on its pKa). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| N-Acetyl-D-glucosaminyl-diphosphodolichol | HMDB | | N-[(2S,3R,4R,5S,6R)-4,5-Dihydroxy-2-{[hydroxy({[hydroxy({[(6E)-3,7,11-trimethyldodeca-6,10-dien-1-yl]oxy})phosphoryl]oxy})phosphoryl]oxy}-6-(hydroxymethyl)oxan-3-yl]ethanimidate | HMDB |
|
|---|
| Chemical Formula | C23H43NO12P2 |
|---|
| Average Molecular Weight | 587.5345 |
|---|
| Monoisotopic Molecular Weight | 587.226048869 |
|---|
| IUPAC Name | {[(2S,3R,4R,5S,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}({[hydroxy({[(6E)-3,7,11-trimethyldodeca-6,10-dien-1-yl]oxy})phosphoryl]oxy})phosphinic acid |
|---|
| Traditional Name | [(2S,3R,4R,5S,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy({hydroxy[(6E)-3,7,11-trimethyldodeca-6,10-dien-1-yl]oxyphosphoryl}oxy)phosphinic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | OC[C@H]1O[C@@H](OP(O)(=O)OP(O)(=O)OCCC(C)CC\C=C(/C)CCC=C(C)C)[C@H](NC(=O)C)[C@@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C23H43NO12P2/c1-15(2)8-6-9-16(3)10-7-11-17(4)12-13-33-37(29,30)36-38(31,32)35-23-20(24-18(5)26)22(28)21(27)19(14-25)34-23/h8,10,17,19-23,25,27-28H,6-7,9,11-14H2,1-5H3,(H,24,26)(H,29,30)(H,31,32)/b16-10+/t17?,19-,20-,21-,22-,23+/m1/s1 |
|---|
| InChI Key | OZGJPDIBINDRGZ-GKXPKBPWSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as n-acyl-alpha-hexosamines. These are carbohydrate derivatives containing a hexose moiety in which the oxygen atom is replaced by an n-acyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | N-acyl-alpha-hexosamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-acyl-alpha-hexosamine
- Farsesane sesquiterpenoid
- Sesquiterpenoid
- Hexose monosaccharide
- Monosaccharide phosphate
- Organic pyrophosphate
- Monoalkyl phosphate
- Monosaccharide
- Organic phosphoric acid derivative
- Alkyl phosphate
- Phosphoric acid ester
- Oxane
- Acetamide
- Carboxamide group
- Secondary carboxylic acid amide
- Secondary alcohol
- Carboxylic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Alcohol
- Organic nitrogen compound
- Carbonyl group
- Primary alcohol
- Organonitrogen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00ai-7809080000-1d59a7a76b20f6c81771 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0036-8829346000-b21da2099e1622d707d1 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("N-Acetyl-D-glucosaminyldiphosphodolichol,1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0019-9316230000-0f0ed3fc6a3e0966260d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-08fu-9617010000-0caba05e81e7f8d9c4c9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-5910000000-0ddc418e7fce8b99a44e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000090000-66563e1b94207dd819a8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0001190000-7a832b88b8ea70dc1e8c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0690-6609100000-8eeff29753a13cc95792 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ab9-0193080000-9443ac239db9306981be | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-2291000000-d8e47b745e7577d5f718 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-4960230000-74dd39082e1fcbef4933 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-1000090000-203baad22ffb155ab510 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00ai-9002520000-a8ba0b10ddee7d955f0a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0k96-9331000000-2e9ed3ec63716d93ea99 | View in MoNA |
|---|
|
|---|