| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:46:16 UTC |
|---|
| Update Date | 2020-04-22 15:08:17 UTC |
|---|
| BMDB ID | BMDB0001458 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Biotin amide |
|---|
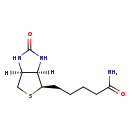
| Description | Biotin amide belongs to the class of organic compounds known as biotin and derivatives. These are organic compounds containing a ureido (tetrahydroimidizalone) ring fused with a tetrahydrothiophene ring. Based on a literature review very few articles have been published on Biotin amide. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 5-[(3AS,6R,6ar)-2-hydroxy-1H,3ah,4H,6H,6ah-thieno[3,4-D]imidazol-6-yl]pentanimidate | Generator, HMDB |
|
|---|
| Chemical Formula | C10H17N3O2S |
|---|
| Average Molecular Weight | 243.326 |
|---|
| Monoisotopic Molecular Weight | 243.104147493 |
|---|
| IUPAC Name | 5-[(3aR,4R,6aS)-2-oxo-hexahydro-1H-thieno[3,4-d]imidazolidin-4-yl]pentanamide |
|---|
| Traditional Name | 5-[(3aR,4R,6aS)-2-oxo-hexahydrothieno[3,4-d]imidazolidin-4-yl]pentanamide |
|---|
| CAS Registry Number | 6929-42-6 |
|---|
| SMILES | [H][C@@]12CS[C@H](CCCCC(N)=O)[C@]1([H])NC(=O)N2 |
|---|
| InChI Identifier | InChI=1S/C10H17N3O2S/c11-8(14)4-2-1-3-7-9-6(5-16-7)12-10(15)13-9/h6-7,9H,1-5H2,(H2,11,14)(H2,12,13,15)/t6-,7-,9-/m1/s1 |
|---|
| InChI Key | XFLVBMBRLSCJAI-ZXFLCMHBSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as biotin and derivatives. These are organic compounds containing a ureido (tetrahydroimidizalone) ring fused with a tetrahydrothiophene ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Biotin and derivatives |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Biotin and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Biotin_derivative
- Thienoimidazolidine
- Fatty amide
- Imidazolidinone
- Fatty acyl
- Thiophene
- Thiolane
- Imidazolidine
- Carboxamide group
- Urea
- Primary carboxylic acid amide
- Carbonic acid derivative
- Azacycle
- Dialkylthioether
- Thioether
- Carboxylic acid derivative
- Organopnictogen compound
- Organonitrogen compound
- Organic nitrogen compound
- Organooxygen compound
- Organic oxygen compound
- Carbonyl group
- Hydrocarbon derivative
- Organic oxide
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0005-9520000000-51917f0c6418688ca18f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004l-0090000000-ee632e902d02574234fa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-1490000000-e498ef37a7c5e9770642 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053v-9600000000-f999e8de54094b58e165 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0190000000-43c6964de1c7149ffeb3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0007-6950000000-055ec27fc755f5f53db3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-37240324d52248632db0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004l-0090000000-4be63d0dd445aeb87350 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-0190000000-5972c30db84501e53f2f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052b-9710000000-b93562f16b5a93447a6f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0090000000-3c9d51b95e146c9f98a5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-2790000000-d7b050a5a96ddf960159 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9300000000-40d0fc7b0203b8ff8417 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|