| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:46:53 UTC |
|---|
| Update Date | 2020-04-22 15:08:28 UTC |
|---|
| BMDB ID | BMDB0001501 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Guanosine diphosphate adenosine |
|---|
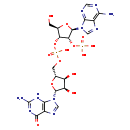
| Description | Guanosine diphosphate adenosine belongs to the class of organic compounds known as (3'->5')-dinucleotides. These are dinucleotides where the two bases are connected via a (3'->5')-phosphodiester linkage. Based on a literature review very few articles have been published on Guanosine diphosphate adenosine. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Guanosine diphosphoric acid adenosine | Generator | | {[(2R,3R,4R,5R)-2-(6-amino-9H-purin-9-yl)-4-[({[(2R,3S,4R,5R)-3,4-dihydroxy-5-(6-hydroxy-2-imino-3,9-dihydro-2H-purin-9-yl)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]-5-(hydroxymethyl)oxolan-3-yl]oxy}phosphonate | HMDB |
|
|---|
| Chemical Formula | C20H26N10O14P2 |
|---|
| Average Molecular Weight | 692.4266 |
|---|
| Monoisotopic Molecular Weight | 692.110518614 |
|---|
| IUPAC Name | {[(2R,3R,4R,5R)-4-[({[(2R,3S,4R,5R)-5-(2-amino-6-oxo-6,9-dihydro-3H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]-2-(6-amino-9H-purin-9-yl)-5-(hydroxymethyl)oxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3R,4R,5R)-4-({[(2R,3S,4R,5R)-5-(2-amino-6-oxo-3H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy)-2-(6-aminopurin-9-yl)-5-(hydroxymethyl)oxolan-3-yl]oxyphosphonic acid |
|---|
| CAS Registry Number | 65331-78-4 |
|---|
| SMILES | NC1=NC(=O)C2=C(N1)N(C=N2)[C@@H]1O[C@H](COP(O)(=O)O[C@@H]2[C@@H](CO)O[C@H]([C@@H]2OP(O)(O)=O)N2C=NC3=C(N)N=CN=C23)[C@@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C20H26N10O14P2/c21-14-8-15(24-3-23-14)29(4-25-8)19-13(43-45(35,36)37)12(6(1-31)41-19)44-46(38,39)40-2-7-10(32)11(33)18(42-7)30-5-26-9-16(30)27-20(22)28-17(9)34/h3-7,10-13,18-19,31-33H,1-2H2,(H,38,39)(H2,21,23,24)(H2,35,36,37)(H3,22,27,28,34)/t6-,7-,10-,11-,12-,13-,18-,19-/m1/s1 |
|---|
| InChI Key | LHSVDZHIRYSVJP-INFSMZHSSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as (3'->5')-dinucleotides. These are dinucleotides where the two bases are connected via a (3'->5')-phosphodiester linkage. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | (3'->5')-dinucleotides and analogues |
|---|
| Sub Class | (3'->5')-dinucleotides |
|---|
| Direct Parent | (3'->5')-dinucleotides |
|---|
| Alternative Parents | |
|---|
| Substituents | - (3'->5')-dinucleotide
- Purine ribonucleoside monophosphate
- Pentose-5-phosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- 6-oxopurine
- Hypoxanthine
- Monosaccharide phosphate
- Purine
- Imidazopyrimidine
- Pyrimidone
- Dialkyl phosphate
- Aminopyrimidine
- Monoalkyl phosphate
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Imidolactam
- Alkyl phosphate
- Pyrimidine
- Vinylogous amide
- Tetrahydrofuran
- Azole
- Heteroaromatic compound
- Imidazole
- Secondary alcohol
- Organoheterocyclic compound
- Oxacycle
- Azacycle
- Amine
- Organic nitrogen compound
- Organic oxygen compound
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Primary alcohol
- Primary amine
- Organopnictogen compound
- Organonitrogen compound
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01r2-8243907000-35e98bb9774530c7cee4 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_16) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f79-0900010000-2517134062e69ae04135 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f79-0900000000-f9331fb92caae2533e9d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f79-0900000000-41b921abfcb64a567415 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0f9x-4901106000-3ea4fd80696ba1393eaa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fai-2900000000-6237eb6ab6c696f039ed | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0059-9800000000-fdac5459a7161cff729b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900014000-60e29d160fae08185d9e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900101000-0a9495b68718f2fd76c3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-0911100000-27a489fb4fc30990db58 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0000009000-ff71e0461774d8b6fea5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05i3-2100209000-8bb9c47b6d7ec520858e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0059-6864219000-89d8fda7e8e5260455bf | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|