| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:47:40 UTC |
|---|
| Update Date | 2020-04-22 15:08:42 UTC |
|---|
| BMDB ID | BMDB0001557 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Riboflavin reduced |
|---|
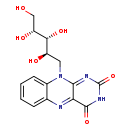
| Description | Riboflavin reduced, also known as reduced riboflavin, belongs to the class of organic compounds known as alloxazines and isoalloxazines. These are organic compounds comprising the (iso)alloxazine structure (Benzo[g]pteridine-2,4-dione). Riboflavin reduced exists in all living organisms, ranging from bacteria to humans. Based on a literature review a small amount of articles have been published on Riboflavin reduced. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 4a,5-Dihydroriboflavin | HMDB | | 4a,5-Dihydroriboflavine | HMDB | | 7,8-Dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)-4a,5-dihydroisoalloxazine | HMDB | | 7,8-Dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)-5,10-dihydrobenzo[g]pteridine-2,4(3H,4ah)-dione | HMDB | | Reduced riboflavin | HMDB |
|
|---|
| Chemical Formula | C15H16N4O6 |
|---|
| Average Molecular Weight | 348.3107 |
|---|
| Monoisotopic Molecular Weight | 348.106984264 |
|---|
| IUPAC Name | 10-[(2R,3R,4R)-2,3,4,5-tetrahydroxypentyl]-2H,3H,4H,10H-benzo[g]pteridine-2,4-dione |
|---|
| Traditional Name | 10-[(2R,3R,4R)-2,3,4,5-tetrahydroxypentyl]-3H-benzo[g]pteridine-2,4-dione |
|---|
| CAS Registry Number | 101652-10-2 |
|---|
| SMILES | OC[C@@H](O)[C@H](O)[C@H](O)CN1C2=CC=CC=C2N=C2C(=O)NC(=O)N=C12 |
|---|
| InChI Identifier | InChI=1S/C15H16N4O6/c20-6-10(22)12(23)9(21)5-19-8-4-2-1-3-7(8)16-11-13(19)17-15(25)18-14(11)24/h1-4,9-10,12,20-23H,5-6H2,(H,18,24,25)/t9-,10-,12-/m1/s1 |
|---|
| InChI Key | ATANIONNQLTUND-CKYFFXLPSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as alloxazines and isoalloxazines. These are organic compounds comprising the (iso)alloxazine structure (Benzo[g]pteridine-2,4-dione). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pteridines and derivatives |
|---|
| Sub Class | Alloxazines and isoalloxazines |
|---|
| Direct Parent | Alloxazines and isoalloxazines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Isoalloxazine
- Diazanaphthalene
- Quinoxaline
- Pyrimidone
- Pyrazine
- Pyrimidine
- Benzenoid
- Heteroaromatic compound
- Vinylogous amide
- Secondary alcohol
- Lactam
- Polyol
- Azacycle
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0bwi-8094000000-f43f7112ee283826153b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-00di-3042269000-7f5ae8c8ce0af8c53f55 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_3_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_4_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_4_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Riboflavin reduced,3TBDMS,#7" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000t-0029000000-bc104c9d050e033b69d4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01t9-4091000000-afc0f707ee1ce75b867c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004i-2190000000-570c6cbcf04a46a68c76 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udl-4097000000-b676ae8b47eab2fb18c7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-f5254591b51a0d846765 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9010000000-0abc9b0d47dd58c290bd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0029000000-01cbdc02a72aeccc6223 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01ta-0098000000-708769959f1af25b7e78 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00b9-0590000000-bc32c00ac0788074d256 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0092000000-f9ec2194483db51c8410 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03fr-1090000000-71092bfa8904438a88ac | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ffx-6590000000-eca89bd048fc24f150a8 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|