| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:49:21 UTC |
|---|
| Update Date | 2020-05-11 20:22:56 UTC |
|---|
| BMDB ID | BMDB0001973 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
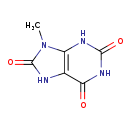
| Common Name | 9-Methyluric acid |
|---|
| Description | 9-Methyluric acid, also known as 9-methylate, belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. Based on a literature review a small amount of articles have been published on 9-Methyluric acid. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 9-Methylate | Generator | | 9-Methylic acid | Generator | | 7,9-dihydro-9-Methyl-1H-purine-2,6,8(3H)-trione | HMDB | | 9-Methyl-2,6,8-trihydroxypurine | HMDB | | MUA | HMDB | | 9-Methyluric acid | MeSH | | 9-Methyl urate | Generator, HMDB |
|
|---|
| Chemical Formula | C6H6N4O3 |
|---|
| Average Molecular Weight | 182.1368 |
|---|
| Monoisotopic Molecular Weight | 182.043990078 |
|---|
| IUPAC Name | 9-methyl-2,3,6,7,8,9-hexahydro-1H-purine-2,6,8-trione |
|---|
| Traditional Name | 9-methyl uric acid |
|---|
| CAS Registry Number | 55441-71-9 |
|---|
| SMILES | CN1C(=O)NC2=C1NC(=O)NC2=O |
|---|
| InChI Identifier | InChI=1S/C6H6N4O3/c1-10-3-2(7-6(10)13)4(11)9-5(12)8-3/h1H3,(H,7,13)(H2,8,9,11,12) |
|---|
| InChI Key | XJEJWDFDVPDMAS-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Xanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthine
- 6-oxopurine
- Purinone
- Alkaloid or derivatives
- Pyrimidone
- N-substituted imidazole
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Vinylogous amide
- Lactam
- Urea
- Azacycle
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 0.021 mg/mL at 25 °C | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0gwr-1900000000-ad4ab961fb153ec968f0 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-001i-0900000000-03ab1b745dbb16bf82cf | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-05o0-4900000000-37b0feef423309da3c70 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0gb9-9800000000-967f7da2548d78a94b57 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0900000000-9cb11ef3bf1ef22898e8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001l-0900000000-2f1b0c821176367c5f41 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0aor-9700000000-06fd51df30b167ff1cf9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0900000000-fc29b0fd4d9a83e9e06c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9500000000-62d351e045a1e54e9a7f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9200000000-15682730273e8e61333b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0900000000-fa33d1f90dc5e1d6cf64 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0900000000-c75e3994fddbcfa2cd2e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0bt9-9700000000-a1642fde92db4b9ec461 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0900000000-35d70442060a47f4840b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fl3-1900000000-05cdb031cf04c4eb174e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9200000000-a0cec5036ddb470e74d7 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, 100%_DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 100%_DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|