| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:50:22 UTC |
|---|
| Update Date | 2020-05-07 14:44:57 UTC |
|---|
| BMDB ID | BMDB0002044 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
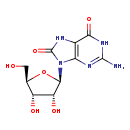
| Common Name | 8-Hydroxyguanosine |
|---|
| Description | 8-Hydroxyguanosine belongs to the class of organic compounds known as purine nucleosides. Purine nucleosides are compounds comprising a purine base attached to a ribosyl or deoxyribosyl moiety. Based on a literature review a significant number of articles have been published on 8-Hydroxyguanosine. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 7,8-dihydro-8-oxo-Guanosine | HMDB | | 8-Oxoguanosine | HMDB, MeSH | | Purine-6,8(1H,9H)-dione, 2-amino-9-.beta.-D-ribofuranosyl- (7ci,8ci) | HMDB | | Purine-6,8(1H,9H)-dione, 2-amino-9-.beta.-delta-ribofuranosyl- (7ci,8ci) | HMDB | | 7,8-dihydro-8-Oxoguanosine | MeSH, HMDB | | 8-OxoGuo | MeSH, HMDB | | 8-oxo-7,8-Dihydroguanosine | MeSH, HMDB | | 8-OxoG nucleoside | MeSH, HMDB | | 8-Hydroxyguanosine | MeSH |
|
|---|
| Chemical Formula | C10H13N5O6 |
|---|
| Average Molecular Weight | 299.2401 |
|---|
| Monoisotopic Molecular Weight | 299.086583173 |
|---|
| IUPAC Name | 2-amino-9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-6,7,8,9-tetrahydro-1H-purine-6,8-dione |
|---|
| Traditional Name | 2-amino-9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,7-dihydropurine-6,8-dione |
|---|
| CAS Registry Number | 3868-31-3 |
|---|
| SMILES | NC1=NC2=C(NC(=O)N2[C@@H]2O[C@H](CO)[C@@H](O)[C@H]2O)C(=O)N1 |
|---|
| InChI Identifier | InChI=1S/C10H13N5O6/c11-9-13-6-3(7(19)14-9)12-10(20)15(6)8-5(18)4(17)2(1-16)21-8/h2,4-5,8,16-18H,1H2,(H,12,20)(H3,11,13,14,19)/t2-,4-,5-,8-/m1/s1 |
|---|
| InChI Key | FPGSEBKFEJEOSA-UMMCILCDSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine nucleosides. Purine nucleosides are compounds comprising a purine base attached to a ribosyl or deoxyribosyl moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleosides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Purine nucleosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine nucleoside
- Glycosyl compound
- N-glycosyl compound
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Hydroxypyrimidine
- Monosaccharide
- N-substituted imidazole
- Pyrimidine
- Heteroaromatic compound
- Azole
- Imidazole
- Tetrahydrofuran
- Secondary alcohol
- Azacycle
- Oxacycle
- Organoheterocyclic compound
- Primary alcohol
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Hydrocarbon derivative
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ab9-9160000000-b5d214bf9c0982dbbe63 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0uec-6902330000-d183539e2e438b4d7c80 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0900000000-0f5c81fc8d1ea968575d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0900000000-0cbf4ce2e5de074bf971 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0v6r-0900000000-625b8c3fb826dde8502a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00kb-0490000000-d4f85e8ee7d5857f5de9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0920000000-8df43d9931a862018046 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00dm-3900000000-5f141ad8cdd5265bd2e2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00kb-0590000000-1ad5ee29954e5c959115 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0930000000-d9cf1a8f55c39625b8e2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00xs-1900000000-6be5e18238989f501180 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gb9-0908000000-df8fac03f541fd5e932a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0900000000-1b32e9ca2d9b8052067d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kb-1910000000-36cbcf5ae3cd08917674 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|