| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:50:34 UTC |
|---|
| Update Date | 2020-04-22 15:09:35 UTC |
|---|
| BMDB ID | BMDB0002060 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
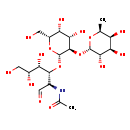
| Common Name | O-6-deoxy-a-L-galactopyranosyl-(1->2)-O-b-D-galactopyranosyl-(1->3)-2-(acetylamino)-2-deoxy-D-Galactose |
|---|
| Description | O-6-deoxy-a-L-galactopyranosyl-(1->2)-O-b-D-galactopyranosyl-(1->3)-2-(acetylamino)-2-deoxy-D-Galactose belongs to the class of organic compounds known as fatty acyl glycosides of mono- and disaccharides. Fatty acyl glycosides of mono- and disaccharides are compounds composed of a mono- or disaccharide moiety linked to one hydroxyl group of a fatty alcohol or of a phosphorylated alcohol (phosphoprenols), a hydroxy fatty acid or to one carboxyl group of a fatty acid (ester linkage) or to an amino alcohol. Based on a literature review a small amount of articles have been published on O-6-deoxy-a-L-galactopyranosyl-(1->2)-O-b-D-galactopyranosyl-(1->3)-2-(acetylamino)-2-deoxy-D-Galactose. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| N-[(2R,3R,4S,5R)-3-{[(2R,3R,4S,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-{[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxy}oxan-2-yl]oxy}-4,5,6-trihydroxy-1-oxohexan-2-yl]ethanimidate | Generator, HMDB | | O-6-Deoxy-a-L-galactopyranosyl-(1->2)-O-b-D-galactopyranosyl-(1->3)-2-(acetylamino)-2-deoxy-D-galactose | Generator | | O-6-Deoxy-α-L-galactopyranosyl-(1->2)-O-β-D-galactopyranosyl-(1->3)-2-(acetylamino)-2-deoxy-D-galactose | Generator, HMDB |
|
|---|
| Chemical Formula | C20H35NO15 |
|---|
| Average Molecular Weight | 529.4896 |
|---|
| Monoisotopic Molecular Weight | 529.200669455 |
|---|
| IUPAC Name | N-[(2R,3R,4S,5R)-3-{[(2R,3R,4S,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-{[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxy}oxan-2-yl]oxy}-4,5,6-trihydroxy-1-oxohexan-2-yl]acetamide |
|---|
| Traditional Name | N-[(2R,3R,4S,5R)-3-{[(2R,3R,4S,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-{[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxy}oxan-2-yl]oxy}-4,5,6-trihydroxy-1-oxohexan-2-yl]acetamide |
|---|
| CAS Registry Number | 95632-88-5 |
|---|
| SMILES | C[C@@H]1O[C@@H](O[C@@H]2[C@@H](O)[C@@H](O)[C@@H](CO)O[C@H]2O[C@@H]([C@@H](O)[C@H](O)CO)[C@@H](NC(C)=O)C=O)[C@@H](O)[C@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C20H35NO15/c1-6-11(27)14(30)16(32)19(33-6)36-18-15(31)13(29)10(5-24)34-20(18)35-17(12(28)9(26)4-23)8(3-22)21-7(2)25/h3,6,8-20,23-24,26-32H,4-5H2,1-2H3,(H,21,25)/t6-,8-,9+,10+,11+,12-,13-,14+,15-,16-,17+,18+,19-,20-/m0/s1 |
|---|
| InChI Key | SJHVKKOMMHUSLU-BKFPXMEMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as fatty acyl glycosides of mono- and disaccharides. Fatty acyl glycosides of mono- and disaccharides are compounds composed of a mono- or disaccharide moiety linked to one hydroxyl group of a fatty alcohol or of a phosphorylated alcohol (phosphoprenols), a hydroxy fatty acid or to one carboxyl group of a fatty acid (ester linkage) or to an amino alcohol. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl glycosides |
|---|
| Direct Parent | Fatty acyl glycosides of mono- and disaccharides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty acyl glycoside of mono- or disaccharide
- Alkyl glycoside
- Disaccharide
- Glycosyl compound
- O-glycosyl compound
- Amino saccharide
- Oxane
- Acetamide
- Secondary carboxylic acid amide
- Secondary alcohol
- Carboxamide group
- Acetal
- Organoheterocyclic compound
- Polyol
- Oxacycle
- Carboxylic acid derivative
- Primary alcohol
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organonitrogen compound
- Organooxygen compound
- Carbonyl group
- Organic nitrogen compound
- Aldehyde
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03di-9201640000-55d9c79756c2bdeeb916 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0a59-7802049000-0f8b88c651dc08e1dfb7 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0hu0-1289250000-f7c5fb7cf2a8a8deee00 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-7986000000-15dfc771f0f21585eba4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ir0-9773000000-0e30805b9e3414713b21 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-7529860000-277fdc2919bcee6c44bf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-7934210000-163ffa8caf3776c2ffe1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-074i-9630000000-ae7728fe66ab853fa706 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-0224290000-2f16468402c071ca384e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-015c-1659000000-e265c17468e375f88c8e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-029j-9400100000-54b72feffe39b638518e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0m29-7312970000-62bc6b0499872f8356e2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9000400000-f148cd7824eec82ac0be | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-8900100000-ee7c820cb9a89dd993ae | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|