| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:53:34 UTC |
|---|
| Update Date | 2020-04-22 15:10:30 UTC |
|---|
| BMDB ID | BMDB0002282 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
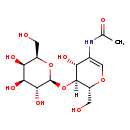
| Common Name | 2-(acetylamino)-1,5-anhydro-2-deoxy-4-O-b-D-galactopyranosyl-D-arabino-Hex-1-enitol |
|---|
| Description | 2-(acetylamino)-1,5-anhydro-2-deoxy-4-O-b-D-galactopyranosyl-D-arabino-Hex-1-enitol belongs to the class of organic compounds known as o-glycosyl compounds. These are glycoside in which a sugar group is bonded through one carbon to another group via a O-glycosidic bond. Based on a literature review very few articles have been published on 2-(acetylamino)-1,5-anhydro-2-deoxy-4-O-b-D-galactopyranosyl-D-arabino-Hex-1-enitol. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| N-[(2R,3S,4R)-4-Hydroxy-2-(hydroxymethyl)-3-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-3,4-dihydro-2H-pyran-5-yl]ethanimidate | HMDB |
|
|---|
| Chemical Formula | C14H23NO10 |
|---|
| Average Molecular Weight | 365.3331 |
|---|
| Monoisotopic Molecular Weight | 365.132195961 |
|---|
| IUPAC Name | N-[(2R,3S,4R)-4-hydroxy-2-(hydroxymethyl)-3-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-3,4-dihydro-2H-pyran-5-yl]acetamide |
|---|
| Traditional Name | N-[(4R,5S,6R)-4-hydroxy-6-(hydroxymethyl)-5-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-5,6-dihydro-4H-pyran-3-yl]acetamide |
|---|
| CAS Registry Number | 868264-13-5 |
|---|
| SMILES | [H][C@@]1(O[C@@H]2O[C@H](CO)[C@H](O)[C@H](O)[C@H]2O)[C@@H](CO)OC=C(NC(C)=O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C14H23NO10/c1-5(18)15-6-4-23-8(3-17)13(9(6)19)25-14-12(22)11(21)10(20)7(2-16)24-14/h4,7-14,16-17,19-22H,2-3H2,1H3,(H,15,18)/t7-,8-,9-,10+,11+,12-,13-,14+/m1/s1 |
|---|
| InChI Key | MIDRXTGJGROJRO-JFXDCXPESA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as o-glycosyl compounds. These are glycoside in which a sugar group is bonded through one carbon to another group via a O-glycosidic bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | O-glycosyl compounds |
|---|
| Alternative Parents | |
|---|
| Substituents | - O-glycosyl compound
- Monosaccharide
- Oxane
- Acetamide
- Carboxamide group
- Secondary alcohol
- Secondary carboxylic acid amide
- Acetal
- Carboxylic acid derivative
- Polyol
- Oxacycle
- Organoheterocyclic compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Alcohol
- Hydrocarbon derivative
- Primary alcohol
- Carbonyl group
- Organic nitrogen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-053b-7559000000-cabc4b335a5da88b1fbe | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-000i-3321039000-fad6fdaa6fdee69926b4 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0uy1-0459000000-a84e893c4fdd72bf4116 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0gw0-0931000000-ed9e33abf55f0740e81b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dl-7910000000-3dc27ddcc09340871b1e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0w29-2469000000-105833db5f04ea0c1b11 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0w29-5944000000-5affc3777ac0c30ed44d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-115c-8910000000-4afd172f4c0d2ac674d0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0009000000-c0a3f459bd00c6946109 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0r00-4922000000-3656fb29090f1318a665 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pc0-9600000000-ea2c55cd4623464e0438 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gw0-0931000000-eedd994a7eeb5fef8873 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-1910000000-f8a62253341bc4ea9aaa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0007-8930000000-66203ec8a95b37f3de43 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|