| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:01:02 UTC |
|---|
| Update Date | 2020-05-11 20:10:18 UTC |
|---|
| BMDB ID | BMDB0002640 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
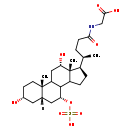
| Common Name | N-[(3a,5b,7a,12a)-3,12-dihydroxy-24-oxo-7-(sulfooxy)cholan-24-yl]-Glycine |
|---|
| Description | N-[(3a,5b,7a,12a)-3,12-dihydroxy-24-oxo-7-(sulfooxy)cholan-24-yl]-Glycine belongs to the class of organic compounds known as glycinated bile acids and derivatives. Glycinated bile acids and derivatives are compounds with a structure characterized by the presence of a glycine linked to a bile acid skeleton. Based on a literature review very few articles have been published on N-[(3a,5b,7a,12a)-3,12-dihydroxy-24-oxo-7-(sulfooxy)cholan-24-yl]-Glycine. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| N-[(3a,5b,7a,12a)-3,12-Dihydroxy-24-oxo-7-(sulphooxy)cholan-24-yl]-glycine | Generator | | 2-{[(4R)-4-[(2S,5R,7S,9R,14R,15R,16S)-5,16-dihydroxy-2,15-dimethyl-9-(sulfooxy)tetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]-1-hydroxypentylidene]amino}acetate | Generator, HMDB | | 2-{[(4R)-4-[(2S,5R,7S,9R,14R,15R,16S)-5,16-dihydroxy-2,15-dimethyl-9-(sulphooxy)tetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]-1-hydroxypentylidene]amino}acetate | Generator, HMDB | | 2-{[(4R)-4-[(2S,5R,7S,9R,14R,15R,16S)-5,16-dihydroxy-2,15-dimethyl-9-(sulphooxy)tetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]-1-hydroxypentylidene]amino}acetic acid | Generator, HMDB |
|
|---|
| Chemical Formula | C26H43NO9S |
|---|
| Average Molecular Weight | 545.686 |
|---|
| Monoisotopic Molecular Weight | 545.265852669 |
|---|
| IUPAC Name | 2-[(4R)-4-[(2S,5R,7S,9R,14R,15R,16S)-5,16-dihydroxy-2,15-dimethyl-9-(sulfooxy)tetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanamido]acetic acid |
|---|
| Traditional Name | [(4R)-4-[(2S,5R,7S,9R,14R,15R,16S)-5,16-dihydroxy-2,15-dimethyl-9-(sulfooxy)tetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanamido]acetic acid |
|---|
| CAS Registry Number | 71977-73-6 |
|---|
| SMILES | [H][C@@]12C[C@H](O)CC[C@]1(C)C1C[C@H](O)[C@]3(C)[C@H](CCC3C1[C@@H](C2)OS(O)(=O)=O)[C@H](C)CCC(=O)NCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C26H43NO9S/c1-14(4-7-22(30)27-13-23(31)32)17-5-6-18-24-19(12-21(29)26(17,18)3)25(2)9-8-16(28)10-15(25)11-20(24)36-37(33,34)35/h14-21,24,28-29H,4-13H2,1-3H3,(H,27,30)(H,31,32)(H,33,34,35)/t14-,15+,16-,17-,18?,19?,20-,21+,24?,25+,26-/m1/s1 |
|---|
| InChI Key | BJPPZTDOLXZUSK-NLKBZSSJSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycinated bile acids and derivatives. Glycinated bile acids and derivatives are compounds with a structure characterized by the presence of a glycine linked to a bile acid skeleton. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Glycinated bile acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycinated bile acid
- Dihydroxy bile acid, alcohol, or derivatives
- Hydroxy bile acid, alcohol, or derivatives
- Sulfated steroid skeleton
- 3-hydroxysteroid
- 12-hydroxysteroid
- Hydroxysteroid
- 3-alpha-hydroxysteroid
- N-acyl-alpha amino acid or derivatives
- N-acyl-alpha-amino acid
- Alpha-amino acid or derivatives
- Sulfate-ester
- Sulfuric acid monoester
- Fatty acyl
- Fatty amide
- Sulfuric acid ester
- Alkyl sulfate
- N-acyl-amine
- Cyclic alcohol
- Organic sulfuric acid or derivatives
- Secondary carboxylic acid amide
- Secondary alcohol
- Carboxamide group
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Alcohol
- Carbonyl group
- Organonitrogen compound
- Organooxygen compound
- Organic oxide
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0gl9-1231490000-253283480a4fc4a47a7b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00di-2110129000-a9dfafcc5584b8686baf | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("N-[(3a,5b,7a,12a)-3,12-dihydroxy-24-oxo-7-(sulfooxy)cholan-24-yl]-Glycine,1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-2000490000-2e8d2cfb1348e6d760d3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0059-6000920000-cca4d584b697cef683eb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004i-9002400000-b0343d0969f16bb3a4f6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002f-0000290000-f5afc42d6a3480129a02 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-3000930000-f62f0db1db3c100bce9d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-9101100000-fbc092ad5516e9401161 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0000090000-fee96f51477bba0d1d44 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-002f-2000290000-1a2b57c158397d0e4193 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-9000320000-8a28de6f794a57ce1524 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004j-0000090000-6b6a2f9cd994ee7fd4bb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03fv-6413490000-b5b0edeb8a2748d05391 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4j-9301100000-bfb0977d7351d65bda46 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|