| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:04:20 UTC |
|---|
| Update Date | 2020-05-19 22:01:35 UTC |
|---|
| BMDB ID | BMDB0003406 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
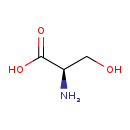
| Common Name | D-Serine |
|---|
| Description | D-Serine, also known as D-serin or DSN, belongs to the class of organic compounds known as serine and derivatives. Serine and derivatives are compounds containing serine or a derivative thereof resulting from reaction of serine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. D-Serine is a very strong basic compound (based on its pKa). D-Serine exists in all living species, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2R)-2-Amino-3-hydroxypropanoic acid | ChEBI | | (R)-2-Amino-3-hydroxy-propionic acid | ChEBI | | (R)-2-Amino-3-hydroxypropanoic acid | ChEBI | | D-Serin | ChEBI | | DSN | ChEBI | | (2R)-2-Amino-3-hydroxypropanoate | Generator | | (R)-2-Amino-3-hydroxy-propionate | Generator | | (R)-2-Amino-3-hydroxypropanoate | Generator | | DL-Serine | HMDB | | Serine D-form | HMDB |
|
|---|
| Chemical Formula | C3H7NO3 |
|---|
| Average Molecular Weight | 105.0926 |
|---|
| Monoisotopic Molecular Weight | 105.042593095 |
|---|
| IUPAC Name | (2R)-2-amino-3-hydroxypropanoic acid |
|---|
| Traditional Name | D-serine |
|---|
| CAS Registry Number | 312-84-5 |
|---|
| SMILES | N[C@H](CO)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C3H7NO3/c4-2(1-5)3(6)7/h2,5H,1,4H2,(H,6,7)/t2-/m1/s1 |
|---|
| InChI Key | MTCFGRXMJLQNBG-UWTATZPHSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as serine and derivatives. Serine and derivatives are compounds containing serine or a derivative thereof resulting from reaction of serine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Serine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Serine or derivatives
- Alpha-amino acid
- D-alpha-amino acid
- Beta-hydroxy acid
- Hydroxy acid
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Alcohol
- Primary amine
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Hydrocarbon derivative
- Primary aliphatic amine
- Organic oxide
- Organopnictogen compound
- Carbonyl group
- Organic oxygen compound
- Amine
- Organic nitrogen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 229 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 364 mg/mL at 20 °C | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-06sl-9000000000-033631650075b940e0e2 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0089-9810000000-06fed7f6e260b107d95a | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-03di-9000000000-d33888cff00d0ade0ffe | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-03di-9000000000-f7e50afcb90768dce4a5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0006-9000000000-043951ed5540cce44f4e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-6900000000-ee98a5a83ffbd434740e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0uki-9200000000-fa8d04d6ce99a290d187 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05fu-9000000000-001b18464fb313594dec | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-9100000000-113290360f404e0abfe6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9000000000-a5d4e47e8a548414bcb5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0596-9000000000-45f597b943c27fee51e7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-08g0-9300000000-dab8446816db786db171 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dl-9000000000-1a5aef91299ef3583ca2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-b6cfe52e68c644cacfd7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-9000000000-c00dff763ff1f56fda00 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-9000000000-980e3923ad0861629a12 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-90338875078d415c5036 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 5%_DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|