| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:04:48 UTC |
|---|
| Update Date | 2020-05-11 20:49:19 UTC |
|---|
| BMDB ID | BMDB0003459 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | D-Alanyl-D-alanine |
|---|
| Description | D-Alanyl-D-alanine, also known as D-ala-D-ala or ala-ala, belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. D-Alanyl-D-alanine is possibly soluble (in water) and a very strong basic compound (based on its pKa). D-Alanyl-D-alanine exists in all living organisms, ranging from bacteria to humans. |
|---|
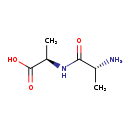
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (D-Ala)2 | ChEBI | | D-Ala-D-ala | ChEBI | | H-D-Ala-D-ala-OH | ChEBI | | Alanyl-D-alanine | HMDB | | Ala-ala | HMDB | | H-Ala-ala-OH | HMDB | | Alanylalanine | HMDB | | Alanylalanine, (D)-isomer | HMDB | | Alanylalanine, (D-ala)-(L-ala)-isomer | HMDB | | Alanylalanine, (L)-isomer | HMDB | | Alanylalanine, (L-ala)-(D-ala)-isomer | HMDB | | Alanylalanine, (L-ala)-(DL-ala)-isomer | HMDB | | Dialanine | HMDB | | D-Alanyl-L-alanine | HMDB | | L-Alanyl-D-alanine | HMDB | | Di-L-alanine | HMDB | | D-Alanylalanine | HMDB | | L-Alanyl-L-alanine | HMDB | | N-D-Alanyl-L-alanine | HMDB | | N-L-Alanyl-D-alanine | HMDB |
|
|---|
| Chemical Formula | C6H12N2O3 |
|---|
| Average Molecular Weight | 160.1711 |
|---|
| Monoisotopic Molecular Weight | 160.08479226 |
|---|
| IUPAC Name | (2R)-2-[(2R)-2-aminopropanamido]propanoic acid |
|---|
| Traditional Name | D-ala-D-ala |
|---|
| CAS Registry Number | 923-16-0 |
|---|
| SMILES | C[C@@H](N)C(=O)N[C@H](C)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H12N2O3/c1-3(7)5(9)8-4(2)6(10)11/h3-4H,7H2,1-2H3,(H,8,9)(H,10,11)/t3-,4-/m1/s1 |
|---|
| InChI Key | DEFJQIDDEAULHB-QWWZWVQMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Dipeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-dipeptide
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- Alpha-amino acid amide
- Alanine or derivatives
- Alpha-amino acid or derivatives
- Amino acid or derivatives
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Primary aliphatic amine
- Organic nitrogen compound
- Carbonyl group
- Primary amine
- Organic oxygen compound
- Hydrocarbon derivative
- Amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0f79-0900000000-5d9afc4a11f868ffd54e | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-014i-0900000000-8c6c90ec8315f93e4ad4 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0f79-0900000000-5d9afc4a11f868ffd54e | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-014i-0900000000-8c6c90ec8315f93e4ad4 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-006x-9100000000-b16775fe354ea161228e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00r6-9400000000-b5b87eed7f3175b80e2e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-03dl-6900000000-90b25f3c22007ea839b5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0a4i-0900000000-bd7876a01b4ebfb8700f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-0006-9000000000-8d6a984538d3c099b09f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0296-5900000000-799966b5c9620715d16a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9200000000-513a3e368e7c6334e54f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006x-9000000000-0724d314ee5219f1c728 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-2900000000-b6d1f12edd1486d8cbb4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-059i-9600000000-8635adeca87ba2d6a666 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00du-9000000000-17647b2edc99cfa2d13b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-052o-4900000000-aae09911d82d8cfafc1f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9100000000-fcc1d4e4f5652338825f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-f5f0d3e84942d47714c6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-9100000000-a9556cdd03ea097d9213 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9000000000-2cbbf675159b82ddf5e8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-ac3751498e9cf095d11f | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| General References | - Pan L, Yu J, Mi Z, Mo L, Jin H, Yao C, Ren D, Menghe B: A Metabolomics Approach Uncovers Differences between Traditional and Commercial Dairy Products in Buryatia (Russian Federation). Molecules. 2018 Mar 22;23(4). pii: molecules23040735. doi: 10.3390/molecules23040735. [PubMed:29565828 ]
|
|---|