| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:04:52 UTC |
|---|
| Update Date | 2020-04-22 15:12:22 UTC |
|---|
| BMDB ID | BMDB0003470 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
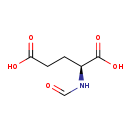
| Common Name | N-Formyl-L-glutamic acid |
|---|
| Description | N-Formyl-L-glutamic acid, also known as N-formyl-L-glutamate, belongs to the class of organic compounds known as glutamic acid and derivatives. Glutamic acid and derivatives are compounds containing glutamic acid or a derivative thereof resulting from reaction of glutamic acid at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. N-Formyl-L-glutamic acid is possibly soluble (in water) and an extremely weak basic (essentially neutral) compound (based on its pKa). N-Formyl-L-glutamic acid exists in all living organisms, ranging from bacteria to humans. Tetrahydrofolic acid and N-formyl-L-glutamic acid can be biosynthesized from N5-formyl-THF and L-glutamic acid; which is mediated by the enzyme formimidoyltransferase-cyclodeaminase. In cattle, N-formyl-L-glutamic acid is involved in the metabolic pathway called the folate metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2-(Formylamino)pentanedioic acid | ChEBI | | N-Formyl-L-glutamate | ChEBI | | (2S)-2-(Formylamino)pentanedioate | Generator | | N-Formylglutamate, ion(2-) | MeSH | | N-Formylglutamate | MeSH | | (2S)-2-Formamidopentanedioate | HMDB | | (2S)-2-Formamidopentanedioic acid | HMDB |
|
|---|
| Chemical Formula | C6H9NO5 |
|---|
| Average Molecular Weight | 175.1394 |
|---|
| Monoisotopic Molecular Weight | 175.048072403 |
|---|
| IUPAC Name | (2S)-2-formamidopentanedioic acid |
|---|
| Traditional Name | N-formyl-L-glutamic acid |
|---|
| CAS Registry Number | 1681-96-5 |
|---|
| SMILES | OC(=O)CC[C@H](NC=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H9NO5/c8-3-7-4(6(11)12)1-2-5(9)10/h3-4H,1-2H2,(H,7,8)(H,9,10)(H,11,12)/t4-/m0/s1 |
|---|
| InChI Key | ADZLWSMFHHHOBV-BYPYZUCNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glutamic acid and derivatives. Glutamic acid and derivatives are compounds containing glutamic acid or a derivative thereof resulting from reaction of glutamic acid at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Glutamic acid and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glutamic acid or derivatives
- N-acyl-l-alpha-amino acid
- N-formyl-alpha-amino acid
- N-formyl-alpha amino acid or derivatives
- Dicarboxylic acid or derivatives
- Fatty acid
- Secondary carboxylic acid amide
- Carboxamide group
- Carboxylic acid
- Organopnictogen compound
- Carbonyl group
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Hydrocarbon derivative
- Organic oxide
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-055f-9500000000-87e21ed84f3e07a0525e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0udi-5090000000-9bdfed547fb095a666a7 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a59-0900000000-56d7b5efa61129bb9f2a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f89-1900000000-f0cf3aae7ee3fc5cdc38 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ue9-9700000000-81fd656d12663befc16e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-05fs-0900000000-e09e113f64d6b1d6f838 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fba-1900000000-df4e50eec3cd34d33f16 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-002f-9300000000-0e7670e199ce4fc65a9a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-053r-0900000000-6fe927d19787d8c02604 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f8i-5900000000-f41841dd571e9833fae3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0084-9000000000-97a1a533c93f062e0524 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00b9-0900000000-db871cdf7055dba8e199 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0f6x-6900000000-421ff60b4e2fe0df43f0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-5a41b69317afca4bfee9 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|