| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:05:00 UTC |
|---|
| Update Date | 2020-05-21 16:29:05 UTC |
|---|
| BMDB ID | BMDB0003514 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Alpha-D-Glucose 1,6-bisphosphate |
|---|
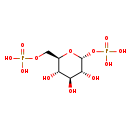
| Description | Alpha-D-Alpha-d-alpha-d-glucose 1,6-bisphosphate, also known as α-Alphalpha-d-alpha-d-glucose 1,6-bisphosphate or alpha-d-glucose 1,6-bisphosphate, belongs to the class of organic compounds known as hexose phosphates. These are carbohydrate derivatives containing a hexose substituted by one or more phosphate groups. Alpha-D-Alpha-d-alpha-d-glucose 1,6-bisphosphate is possibly soluble (in water) and an extremely weak basic (essentially neutral) compound (based on its pKa). Alpha-D-Alpha-d-alpha-d-glucose 1,6-bisphosphate exists in all living species, ranging from bacteria to humans. Alpha-D-Alpha-d-alpha-d-glucose 1,6-bisphosphate and 3-phosphoglyceric acid can be biosynthesized from glucose 1-phosphate and glyceric acid 1,3-biphosphate through the action of the enzyme alpha-d-glucose 1,6-bisphosphate synthase. In cattle, Alpha-Alpha-alphalpha-d-alpha-d-glucose 1,6-bisphosphate is involved in the metabolic pathway called the starch and sucrose metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| alpha-D-Glucose 1,6-biphosphate | ChEBI | | D-Glucose 1,6-biphosphate | ChEBI | | D-Glucose 1,6-bisphosphate | Kegg | | a-D-Glucose 1,6-biphosphate | Generator | | a-D-Glucose 1,6-biphosphoric acid | Generator | | alpha-D-Glucose 1,6-biphosphoric acid | Generator | | Α-D-glucose 1,6-biphosphate | Generator | | Α-D-glucose 1,6-biphosphoric acid | Generator | | D-Glucose 1,6-biphosphoric acid | Generator | | D-Glucose 1,6-bisphosphoric acid | Generator | | a-D-Glucose 1,6-bisphosphate | Generator | | a-D-Glucose 1,6-bisphosphoric acid | Generator | | alpha-D-Glucose 1,6-bisphosphoric acid | Generator | | Α-D-glucose 1,6-bisphosphate | Generator | | Α-D-glucose 1,6-bisphosphoric acid | Generator | | a-D-Glucose 1,6-bis(dihydrogen phosphate) | HMDB | | a-D-Glucose 1,6-diphosphate | HMDB | | alpha-D-1,6-Bis(dihydrogen phosphate) glucopyranose | HMDB | | alpha-D-Glucose 1,6-bis(dihydrogen phosphate) | HMDB | | alpha-D-Glucose 1,6-diphosphate | HMDB | | alpha-delta-1,6-Bis(dihydrogen phosphate) glucopyranose | HMDB | | alpha-delta-Glucose 1,6-bis(dihydrogen phosphate) | HMDB | | alpha-delta-Glucose 1,6-bisphosphate | HMDB | | alpha-delta-Glucose 1,6-diphosphate | HMDB | | D-Glucose 1,6-diphosphate | HMDB | | delta-Glucose 1,6-diphosphate | HMDB | | Glucose 1,6-bisphosphate | HMDB | | Glucose 1,6-diphosphate | HMDB | | beta-D-Glucose 1,6-(bis)phosphate | HMDB | | Glucose-1,6-diphosphate | HMDB | | Glucose-1,6-bisphosphate | HMDB | | alpha-Glucose 1,6-diphosphate | HMDB | | Α-D-glucose 1,6-diphosphate | HMDB | | Α-glucose 1,6-diphosphate | HMDB | | alpha-D-Glucose 1,6-bisphosphate | HMDB |
|
|---|
| Chemical Formula | C6H14O12P2 |
|---|
| Average Molecular Weight | 340.1157 |
|---|
| Monoisotopic Molecular Weight | 339.996048936 |
|---|
| IUPAC Name | {[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-[(phosphonooxy)methyl]oxan-2-yl]oxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-[(phosphonooxy)methyl]oxan-2-yl]oxyphosphonic acid |
|---|
| CAS Registry Number | 10139-18-1 |
|---|
| SMILES | O[C@H]1[C@H](O)[C@@H](COP(O)(O)=O)O[C@H](OP(O)(O)=O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C6H14O12P2/c7-3-2(1-16-19(10,11)12)17-6(5(9)4(3)8)18-20(13,14)15/h2-9H,1H2,(H2,10,11,12)(H2,13,14,15)/t2-,3-,4+,5-,6-/m1/s1 |
|---|
| InChI Key | RWHOZGRAXYWRNX-VFUOTHLCSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hexose phosphates. These are carbohydrate derivatives containing a hexose substituted by one or more phosphate groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Hexose phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hexose phosphate
- Monosaccharide phosphate
- Monoalkyl phosphate
- Alkyl phosphate
- Phosphoric acid ester
- Oxane
- Organic phosphoric acid derivative
- Secondary alcohol
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Organic oxide
- Hydrocarbon derivative
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | -4.374 | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9422000000-7bfff1b0b2d51cca5dff | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0fr5-7922130000-d6ba2057c5f43a10b976 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-002f-9182000000-23bca820b90a45f17acb | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 23V, positive | splash10-00dl-0089000000-14b2732dbc26e1c46650 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 23V, positive | splash10-03di-0090000000-eff1032954c312ffe781 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 4V, positive | splash10-03di-0019000000-b6e1680b5b949ae7c8f6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 6V, positive | splash10-03xr-0069000000-63772b51257cee2af15a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 9V, positive | splash10-014i-0092000000-65fab63b500e60a0ea01 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 14V, positive | splash10-014i-0190000000-bfbcf573cf5f756927f9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 19V, positive | splash10-01b9-0790000000-757dcf8e67df0907f65a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 24V, positive | splash10-00di-0920000000-18f7c04ce401fda68354 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 23V, positive | splash10-014i-0090000000-e9698a8c472de2ec65b3 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 23V, positive | splash10-00fr-0930000000-7f0cb15d50e589a9640f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 23V, positive | splash10-0002-0910000000-4a92067b100f48729122 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 23V, positive | splash10-0002-0390000000-c8d4b6af4a7e1dbc6766 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 23V, positive | splash10-004i-0069000000-b2d82f85ee47b9928a9c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 23V, positive | splash10-004i-0091000000-e9e211cdeab0d7aab3ab | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 4V, positive | splash10-014i-0000900000-386775c642a8d4f6b7b3 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 7V, positive | splash10-014i-0004900000-249416017fe86caa0eb1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 10V, positive | splash10-014i-0109200000-91fd1405fb3d3ce23e3d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 16V, positive | splash10-014i-0309000000-71838f11ad810bfe78ac | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-2169000000-7496db21481f5c6646f1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0007-8194000000-0ab11c338e7462b1784e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9710000000-04febc887536086e1a99 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002r-7319000000-8f0f5f6bb5550e944766 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-e67858a197b35311ba75 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-44fd1ff1d686ff20aa4f | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|