| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:18:35 UTC |
|---|
| Update Date | 2020-04-22 15:16:20 UTC |
|---|
| BMDB ID | BMDB0005794 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Quercetin |
|---|
| Description | Quercetin, also known as sophoretin or xanthaurine, belongs to the class of organic compounds known as flavonols. Flavonols are compounds that contain a flavone (2-phenyl-1-benzopyran-4-one) backbone carrying a hydroxyl group at the 3-position. Thus, quercetin is considered to be a flavonoid. Quercetin exists in all living species, ranging from bacteria to plants to humans. Based on a literature review a significant number of articles have been published on Quercetin. |
|---|
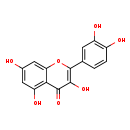
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-(3,4-Dihydroxyphenyl)-3,5,7-trihydroxy-4H-1-benzopyran-4-one | ChEBI | | 3,3',4',5,7-Pentahydroxyflavone | ChEBI | | 3,5,7,3',4'-Pentahydroxyflavone | ChEBI | | Sophoretin | ChEBI | | Xanthaurine | ChEBI | | 3,3',4,5,7-Pentahydroxyflavone | Kegg | | Dikvertin | MeSH | | 2-(3,4-Dihydroxy-phenyl)-3,5,7-trihydroxy-chromen-4-one | HMDB | | 3',4',5,7-Tetrahydroxyflavan-3-ol | HMDB | | 3',4',5,7-Tetrahydroxyflavon-3-ol | HMDB | | 3,4',5,5',7-Pentahydroxy-flavone | HMDB | | 3,5,7-Trihydroxy-2-(3,4-dihydroxyphenyl)-4H-chromen-4-ON | HMDB | | Flavin meletin | HMDB | | Meletin | HMDB | | Quercetin dihydrate | HMDB | | Quercetol | HMDB | | Quertin | HMDB | | 2-(3,4-Dihydroxyphenyl)-3,5,7-trihydroxy-4H-benzopyran-4-one | PhytoBank | | 3,3’,4’,5,7-Pentahydroxyflavone | PhytoBank | | 3,5,7,3’,4’-Pentahydroxyflavone | PhytoBank | | 3,5,7-Trihydroxy-2-(3,4-dihydroxyphenyl)-4H-chromen-4-one | PhytoBank | | 3'-Hydroxykaempferol | PhytoBank | | 3’-Hydroxykaempferol | PhytoBank | | Quercetine | PhytoBank | | Quertine | PhytoBank |
|
|---|
| Chemical Formula | C15H10O7 |
|---|
| Average Molecular Weight | 302.2357 |
|---|
| Monoisotopic Molecular Weight | 302.042652674 |
|---|
| IUPAC Name | 2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-one |
|---|
| Traditional Name | quercetin |
|---|
| CAS Registry Number | 117-39-5 |
|---|
| SMILES | OC1=CC2=C(C(O)=C1)C(=O)C(O)=C(O2)C1=CC=C(O)C(O)=C1 |
|---|
| InChI Identifier | InChI=1S/C15H10O7/c16-7-4-10(19)12-11(5-7)22-15(14(21)13(12)20)6-1-2-8(17)9(18)3-6/h1-5,16-19,21H |
|---|
| InChI Key | REFJWTPEDVJJIY-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as flavonols. Flavonols are compounds that contain a flavone (2-phenyl-1-benzopyran-4-one) backbone carrying a hydroxyl group at the 3-position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Phenylpropanoids and polyketides |
|---|
| Class | Flavonoids |
|---|
| Sub Class | Flavones |
|---|
| Direct Parent | Flavonols |
|---|
| Alternative Parents | |
|---|
| Substituents | - 3-hydroxyflavone
- 3'-hydroxyflavonoid
- 3-hydroxyflavonoid
- 4'-hydroxyflavonoid
- 5-hydroxyflavonoid
- 7-hydroxyflavonoid
- Hydroxyflavonoid
- Chromone
- Benzopyran
- 1-benzopyran
- Catechol
- 1-hydroxy-4-unsubstituted benzenoid
- 1-hydroxy-2-unsubstituted benzenoid
- Phenol
- Pyranone
- Benzenoid
- Monocyclic benzene moiety
- Pyran
- Heteroaromatic compound
- Vinylogous acid
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Organic oxide
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 316 - 318 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 0.06 mg/mL at 16 °C | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (5 TMS) | splash10-0bt9-2611390000-f8e98c928a7ed82acda4 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-0bt9-2611390000-f8e98c928a7ed82acda4 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0079-0591000000-2a146657da898ec9322e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (5 TMS) - 70eV, Positive | splash10-00l2-2093078000-1de46637305246feffd2 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0udi-0009000000-d4689b76f41c73487399 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0udi-0967000000-613e61ec0c69ed0ee630 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0uy0-5910000000-ee816015eec26c8621b1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) 5V, Positive | splash10-0udi-0009000000-ec1cab852ed9f9f78fa4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0udi-1907000000-6f36df2733dadae380c2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0udi-0309000000-976a99c106ceca16d73b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0pi0-1900000000-b2e286366d41e47dd8fc | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0udi-0209000000-e891863ec110aeb660b0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-0udi-0940000000-aa52db00c1defe3ccf75 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-0udi-0219000000-5ef285c4b6bfd220b8b1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-0udi-0219000000-547c83bb70e7da007d6c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0udi-1907000000-a59602c09f66e9656068 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITTOF (LCMS-IT-TOF) , Positive | splash10-0udi-0009000000-4416f39adf6c9b919bfa | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITTOF (LCMS-IT-TOF) , Negative | splash10-0udi-0019000000-eb14ec62fc2fb2f1da88 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITTOF (LCMS-IT-TOF) , Negative | splash10-0ufr-0910000000-0730bca525c17aac75c5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF 10V, Negative | splash10-004i-0030290000-06238e4a98a4daad3265 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF , Negative | splash10-0udi-0039008002-9df3edfb34deb15f8474 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF 10V, Negative | splash10-0udi-0039008002-9df3edfb34deb15f8474 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - ESI-TOF 20V, Negative | splash10-0uka-0193000000-3325ca1a080730a9c0bf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0019000000-ee8570ac70818e8939bb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0279000000-71b5db7ef8322cc8b9e9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0zg0-7960000000-7edbb229853a642749ac | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0009000000-21580b9d8394d2eea3a5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0239000000-b77fcb9c0ce11dc515bf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0apr-5940000000-0590bdee504c5ed6ba36 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, 100%_DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 100%_DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|