| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:19:23 UTC |
|---|
| Update Date | 2020-04-22 15:16:35 UTC |
|---|
| BMDB ID | BMDB0005960 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | D-Pipecolic acid |
|---|
| Description | D-Pipecolic acid, also known as 6-carboxypiperidine or (R)-pipecolate, belongs to the class of organic compounds known as d-alpha-amino acids. These are alpha amino acids which have the D-configuration of the alpha-carbon atom. D-Pipecolic acid exists as a solid, possibly soluble (in water), and a very strong basic compound (based on its pKa) molecule. |
|---|
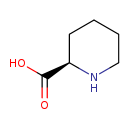
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (R)-Pipecolic acid | ChEBI | | (R)-Piperidine-2-carboxylic acid | ChEBI | | 6-CARBOXYPIPERIDINE | ChEBI | | (R)-Pipecolate | Generator | | (R)-Piperidine-2-carboxylate | Generator | | D-Pipecolate | Generator | | (+)-Pipecolic acid | HMDB | | (+)-Pipecolinic acid | HMDB | | (2R)-2-Piperidinecarboxylic acid | HMDB | | (2R)-Piperidine-2-carboxylic acid | HMDB | | (R)-()-2-Piperidinecarboxylic acid | HMDB | | (R)-(+)-Pipecolic acid | HMDB | | (R)-2-Piperidinecarboxylic acid | HMDB | | (R)-Pipecolinic acid | HMDB | | D-(+)- Pipecolic acid | HMDB | | D-(+)-Pipecolic acid | HMDB | | D-Homoproline | HMDB | | D-Pipecolinic acid | HMDB | | D-Piperidine-2-carboxylic acid | HMDB |
|
|---|
| Chemical Formula | C6H11NO2 |
|---|
| Average Molecular Weight | 129.157 |
|---|
| Monoisotopic Molecular Weight | 129.078978601 |
|---|
| IUPAC Name | (2R)-piperidine-2-carboxylic acid |
|---|
| Traditional Name | (+)-pipecolic acid |
|---|
| CAS Registry Number | 1723-00-8 |
|---|
| SMILES | OC(=O)[C@H]1CCCCN1 |
|---|
| InChI Identifier | InChI=1S/C6H11NO2/c8-6(9)5-3-1-2-4-7-5/h5,7H,1-4H2,(H,8,9)/t5-/m1/s1 |
|---|
| InChI Key | HXEACLLIILLPRG-RXMQYKEDSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as d-alpha-amino acids. These are alpha amino acids which have the D-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | D-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - D-alpha-amino acid
- Piperidinecarboxylic acid
- Piperidine
- Amino acid
- Carboxylic acid
- Secondary aliphatic amine
- Monocarboxylic acid or derivatives
- Secondary amine
- Organoheterocyclic compound
- Azacycle
- Organic oxygen compound
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Amine
- Carbonyl group
- Organopnictogen compound
- Hydrocarbon derivative
- Organic oxide
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 266 - 268 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-001i-9100000000-335cf7f41fca5cbc84cf | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-001i-9100000000-4611bf819d3b9d4fd062 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-3900000000-bf71d20a7c8e71d335e5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9400000000-fa70645b026293d659de | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053u-9000000000-f849cc3c4b11bc7d4de7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-2900000000-bf800b5cd197887a11d5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-003r-7900000000-8a0e9f51df4006740863 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-9000000000-effc196b5c15971f985a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-9300000000-cb8df30f7267d610fd8d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9000000000-72c8691eb15dce6e0abb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053r-9000000000-c7623c5058ad6c879ec3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0900000000-b487cd2a72f17977a8cd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01t9-1900000000-83b59287dc6d3e726326 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0036-9200000000-3ca813e7348e686ed922 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Fadel, Antoine; Lahrache, Nabil. An efficient synthesis of enantiomerically pure (R)-pipecolic acid, (S)-proline, and their N-alkylated derivatives. Journal of Organic Chemistry (2007), 72(5), 1780-1784. |
|---|