| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:23:16 UTC |
|---|
| Update Date | 2020-04-22 15:17:49 UTC |
|---|
| BMDB ID | BMDB0006555 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | dIMP |
|---|
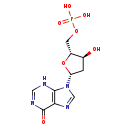
| Description | dIMP, also known as deoxy imp, belongs to the class of organic compounds known as purine 2'-deoxyribonucleoside monophosphates. These are purine nucleotides with monophosphate group linked to the ribose moiety lacking a hydroxyl group at position 2. dIMP is a moderately basic compound (based on its pKa). dIMP exists in all living organisms, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2'-Deoxy-5'-inosinic acid | ChEBI | | 2'-Deoxyinosine 5'-monophosphate | ChEBI | | 2'-Deoxyinosine 5'-monophosphoric acid | ChEBI | | 2'-Deoxyinosine 5'-phosphate | ChEBI | | 9-(2-Deoxy-5-O-phosphono-beta-D-erythro-pentofuranosyl)-9H-purin-6-ol | ChEBI | | 9-(2-Deoxy-5-O-phosphono-beta-delta-erythro-pentofuranosyl)-9H-purin-6-ol | ChEBI | | Deoxyinosine monophosphate | ChEBI | | 2'-Deoxy-5'-inosinate | Generator | | 2'-Deoxyinosine 5'-phosphoric acid | Generator | | 9-(2-Deoxy-5-O-phosphono-b-D-erythro-pentofuranosyl)-9H-purin-6-ol | Generator | | 9-(2-Deoxy-5-O-phosphono-β-D-erythro-pentofuranosyl)-9H-purin-6-ol | Generator | | 9-(2-Deoxy-5-O-phosphono-b-delta-erythro-pentofuranosyl)-9H-purin-6-ol | Generator | | 9-(2-Deoxy-5-O-phosphono-β-δ-erythro-pentofuranosyl)-9H-purin-6-ol | Generator | | Deoxyinosine monophosphoric acid | Generator | | 9-(2-Deoxy-5-O-phosphono-b-δ-erythro-pentofuranosyl)-9H-purin-6-ol | HMDB | | Hypoxanthine deoxyriboside | HMDB | | [(2R,3S,4R,5R)-3-Hydroxy-5-(6-hydroxy-9H-purin-9-yl)tetrahydrofuran-2-yl]methyl dihydrogen phosphate | HMDB | | Deoxy imp | HMDB | | 2'-Deoxyinosine-5'-monophosphoric acid | HMDB | | DIMP | ChEBI |
|
|---|
| Chemical Formula | C10H13N4O7P |
|---|
| Average Molecular Weight | 332.2066 |
|---|
| Monoisotopic Molecular Weight | 332.052185302 |
|---|
| IUPAC Name | {[(2R,3S,5R)-3-hydroxy-5-(6-oxo-6,9-dihydro-3H-purin-9-yl)oxolan-2-yl]methoxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3S,5R)-3-hydroxy-5-(6-oxo-3H-purin-9-yl)oxolan-2-yl]methoxyphosphonic acid |
|---|
| CAS Registry Number | 3393-18-8 |
|---|
| SMILES | O[C@H]1C[C@@H](O[C@@H]1COP(O)(O)=O)N1C=NC2=C1NC=NC2=O |
|---|
| InChI Identifier | InChI=1S/C10H13N4O7P/c15-5-1-7(21-6(5)2-20-22(17,18)19)14-4-13-8-9(14)11-3-12-10(8)16/h3-7,15H,1-2H2,(H,11,12,16)(H2,17,18,19)/t5-,6+,7+/m0/s1 |
|---|
| InChI Key | PHNGFPPXDJJADG-RRKCRQDMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine 2'-deoxyribonucleoside monophosphates. These are purine nucleotides with monophosphate group linked to the ribose moiety lacking a hydroxyl group at position 2. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine deoxyribonucleotides |
|---|
| Direct Parent | Purine 2'-deoxyribonucleoside monophosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine 2'-deoxyribonucleoside monophosphate
- Hypoxanthine
- Imidazopyrimidine
- Purine
- Hydroxypyrimidine
- Monoalkyl phosphate
- N-substituted imidazole
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Pyrimidine
- Alkyl phosphate
- Azole
- Imidazole
- Heteroaromatic compound
- Tetrahydrofuran
- Secondary alcohol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Alcohol
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (4 TMS) | splash10-001i-9210000000-53948c1a90a0f6914597 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-001i-9210000000-53948c1a90a0f6914597 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9401000000-3e05abb6a8d8b1a335b6 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00rx-7923000000-8831273c387905273e72 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-001r-0619000000-1a0eb2cb24a05602189f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) 30V, Positive | splash10-000i-2902000000-8031212ff8d5a3caf932 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-005j-9803000000-e0eccae074a61767e080 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-005j-9803000000-e0eccae074a61767e080 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-001r-0619000000-1a0eb2cb24a05602189f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-000i-2902000000-8031212ff8d5a3caf932 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-000i-2902000000-8031212ff8d5a3caf932 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0901000000-82485bc6f5afd265ffb6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0900000000-40c3e510e6f14b9c6415 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0900000000-084491e4ef779439c6e7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-003r-5209000000-fe97448e2adad18be4e1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004r-9400000000-69f08fb1d6025455e334 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-eaad95f8c8f37f17a06d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0900000000-b2e36fb98660834d7d24 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0900000000-c7e643ad3fc1ffc399bb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0019-5900000000-13459080c7272714095d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-003r-9008000000-2805c5fa43654779dc36 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9101000000-bc2b5d6253fd7cf52a89 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-ee07b6d42071fb27d3f4 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|