| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:23:37 UTC |
|---|
| Update Date | 2020-04-22 15:17:56 UTC |
|---|
| BMDB ID | BMDB0006579 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 3-Sialyl Lewis |
|---|
| Description | 3-Sialyl Lewis belongs to the class of organic compounds known as n-acylneuraminic acids. These are neuraminic acids carrying an N-acyl substituent. Based on a literature review very few articles have been published on 3-Sialyl Lewis. |
|---|
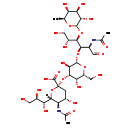
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3'-Sialyl lewis a | HMDB | | O-(N-Acetyl-alpha-neuraminosyl)-(2->3)-O-beta-D-galactopyranosyl-(1->3)-O-[6-deoxy-alpha-L-galactopyranosyl-(1->4)]-2-(acetylamino)-2-deoxy- D-glucose | HMDB | | O-(N-Acetyl-alpha-neuraminosyl)-(2->3)-O-beta-delta-galactopyranosyl-(1->3)-O-[6-deoxy-alpha-L-galactopyranosyl-(1->4)]-2-(acetylamino)-2-deoxy- D-glucose | HMDB | | Sialyl lea tri | HMDB | | Sialyl lewis a | HMDB | | SLea | HMDB | | (2S,4S,5R,6R)-2-{[(2R,3R,4S,5S,6R)-2-{[(2R,3R,4R,5R)-5,6-dihydroxy-2-[(1-hydroxyethylidene)amino]-1-oxo-4-{[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxy}hexan-3-yl]oxy}-3,5-dihydroxy-6-(hydroxymethyl)oxan-4-yl]oxy}-4-hydroxy-5-[(1-hydroxyethylidene)amino]-6-[(1R,2R)-1,2,3-trihydroxypropyl]oxane-2-carboxylate | Generator, HMDB |
|
|---|
| Chemical Formula | C31H52N2O23 |
|---|
| Average Molecular Weight | 820.7442 |
|---|
| Monoisotopic Molecular Weight | 820.29608598 |
|---|
| IUPAC Name | (2S,4S,5R,6R)-5-acetamido-2-{[(2R,3R,4S,5S,6R)-2-{[(2R,3R,4R,5R)-2-acetamido-5,6-dihydroxy-1-oxo-4-{[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxy}hexan-3-yl]oxy}-3,5-dihydroxy-6-(hydroxymethyl)oxan-4-yl]oxy}-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]oxane-2-carboxylic acid |
|---|
| Traditional Name | sialyl-lewis A |
|---|
| CAS Registry Number | 92448-22-1 |
|---|
| SMILES | [H][C@]1(O[C@@](C[C@H](O)[C@H]1NC(C)=O)(O[C@H]1[C@@H](O)[C@@H](CO)O[C@@H](O[C@H]([C@@H](NC(C)=O)C=O)[C@H](O[C@@H]2O[C@@H](C)[C@@H](O)[C@@H](O)[C@@H]2O)[C@H](O)CO)[C@@H]1O)C(O)=O)[C@H](O)[C@H](O)CO |
|---|
| InChI Identifier | InChI=1S/C31H52N2O23/c1-9-18(43)21(46)22(47)28(51-9)54-25(15(42)7-36)24(12(5-34)32-10(2)38)53-29-23(48)27(20(45)16(8-37)52-29)56-31(30(49)50)4-13(40)17(33-11(3)39)26(55-31)19(44)14(41)6-35/h5,9,12-29,35-37,40-48H,4,6-8H2,1-3H3,(H,32,38)(H,33,39)(H,49,50)/t9-,12-,13-,14+,15+,16+,17+,18+,19+,20-,21+,22-,23+,24+,25+,26+,27-,28-,29-,31-/m0/s1 |
|---|
| InChI Key | INZOTETZQBPBCE-NYLDSJSYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as n-acylneuraminic acids. These are neuraminic acids carrying an N-acyl substituent. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | N-acylneuraminic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-acylneuraminic acid
- Neuraminic acid
- Fatty acyl glycoside of mono- or disaccharide
- Fatty acyl glycoside
- C-glucuronide
- Alkyl glycoside
- O-glycosyl compound
- Glycosyl compound
- C-glycosyl compound
- Amino saccharide
- Ketal
- Oxane
- Pyran
- Monosaccharide
- Fatty acyl
- Acetamide
- Secondary carboxylic acid amide
- Secondary alcohol
- Carboxamide group
- Acetal
- Carboxylic acid derivative
- Carboxylic acid
- Oxacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Polyol
- Alcohol
- Hydrocarbon derivative
- Carbonyl group
- Aldehyde
- Organic nitrogen compound
- Primary alcohol
- Organopnictogen compound
- Organic oxide
- Organonitrogen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_16) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0039047780-a6a6eefa31eb1cd8aa37 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-1219050000-24ed19da17cf8095551a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ht9-2197012000-e61172bbfcd93fa7dc71 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0kml-3811003940-bfbedd8c5c1a742e1588 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0pdr-9847130210-b52a4b20d0eb31cc3b79 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-8659000000-a3d562e1e1522405c2b1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0fk9-0002201290-3aa7a8d28cf5c288102b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05gi-0035397460-100165c981288423ab19 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-02ai-9742023200-8f288fed4a7cfc3f3b4a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0le9-6141001890-f11d5c1fbe0826e11e5f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-053r-2160031930-ba7fd20ea109f7ae0aef | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ldi-4297000000-12c687ed5acc9af26505 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|