| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:23:51 UTC |
|---|
| Update Date | 2020-04-22 15:18:00 UTC |
|---|
| BMDB ID | BMDB0006592 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Lacto-N-triaose |
|---|
| Description | Lacto-N-triaose belongs to the class of organic compounds known as oligosaccharides. These are carbohydrates made up of 3 to 10 monosaccharide units linked to each other through glycosidic bonds. Based on a literature review a small amount of articles have been published on Lacto-N-triaose. |
|---|
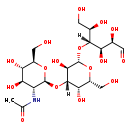
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Agalacto-N-neotetraose | HMDB | | Lacto-N-triose II | HMDB | | O-2-(Acetylamino)-2-deoxy-beta-D-glucopyranosyl-(1->3)-O-beta-D-galactopyranosyl-(1->4)-D-glucose | HMDB | | O-2-(Acetylamino)-2-deoxy-beta-delta-glucopyranosyl-(1->3)-O-beta-delta-galactopyranosyl-(1->4)-delta-glucose | HMDB | | beta-D-GlcNAc(1-3)-beta-D-gal(1-4)-beta-D-GLC-ome | HMDB | | Methyl 2-acetamido-2-deoxyglucopyranosyl-1-3-galactopyranosyl-1-4-glucopyranoside | HMDB | | GlcNAc-1-3-gal-1-4-GLC-ome | HMDB | | N-[(2S,3R,4R,5S,6R)-2-{[(2R,3S,4S,5R,6S)-3,5-dihydroxy-2-(hydroxymethyl)-6-{[(2R,3R,4R,5R)-1,2,4,5-tetrahydroxy-6-oxohexan-3-yl]oxy}oxan-4-yl]oxy}-4,5-dihydroxy-6-(hydroxymethyl)oxan-3-yl]ethanimidate | HMDB |
|
|---|
| Chemical Formula | C20H35NO16 |
|---|
| Average Molecular Weight | 545.489 |
|---|
| Monoisotopic Molecular Weight | 545.195584077 |
|---|
| IUPAC Name | N-[(2S,3R,4R,5S,6R)-2-{[(2R,3S,4S,5R,6S)-3,5-dihydroxy-2-(hydroxymethyl)-6-{[(2R,3R,4R,5R)-1,2,4,5-tetrahydroxy-6-oxohexan-3-yl]oxy}oxan-4-yl]oxy}-4,5-dihydroxy-6-(hydroxymethyl)oxan-3-yl]acetamide |
|---|
| Traditional Name | N-[(2S,3R,4R,5S,6R)-2-{[(2R,3S,4S,5R,6S)-3,5-dihydroxy-2-(hydroxymethyl)-6-{[(2R,3R,4R,5R)-1,2,4,5-tetrahydroxy-6-oxohexan-3-yl]oxy}oxan-4-yl]oxy}-4,5-dihydroxy-6-(hydroxymethyl)oxan-3-yl]acetamide |
|---|
| CAS Registry Number | 75645-27-1 |
|---|
| SMILES | [H][C@@](O[C@@H]1O[C@H](CO)[C@H](O)[C@]([H])(O[C@@H]2O[C@H](CO)[C@@H](O)[C@H](O)[C@H]2NC(C)=O)[C@H]1O)([C@H](O)CO)[C@H](O)[C@@H](O)C=O |
|---|
| InChI Identifier | InChI=1S/C20H35NO16/c1-6(26)21-11-15(32)13(30)9(4-24)34-19(11)37-18-14(31)10(5-25)35-20(16(18)33)36-17(8(28)3-23)12(29)7(27)2-22/h2,7-20,23-25,27-33H,3-5H2,1H3,(H,21,26)/t7-,8+,9+,10+,11+,12+,13+,14-,15+,16+,17+,18-,19-,20-/m0/s1 |
|---|
| InChI Key | RJTOFDPWCJDYFZ-SPVZFZGWSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as oligosaccharides. These are carbohydrates made up of 3 to 10 monosaccharide units linked to each other through glycosidic bonds. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Oligosaccharides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Oligosaccharide
- Fatty acyl glycoside
- N-acyl-alpha-hexosamine
- Alkyl glycoside
- Glycosyl compound
- O-glycosyl compound
- Beta-hydroxy aldehyde
- Fatty acyl
- Oxane
- Acetamide
- Alpha-hydroxyaldehyde
- Carboxamide group
- Secondary carboxylic acid amide
- Secondary alcohol
- Acetal
- Carboxylic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Hydrocarbon derivative
- Alcohol
- Organic oxide
- Carbonyl group
- Organopnictogen compound
- Aldehyde
- Primary alcohol
- Organic nitrogen compound
- Organonitrogen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03di-9112450000-b0c295c0a682d2bbd999 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0089-4914017000-c0d5e971a5f54e7f92ac | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Lacto-N-triaose,1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01u0-1901280000-b64214998b2fdd5a49a0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03gi-5913110000-43a3955a51ad602c9d50 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03e9-8920100000-4bfa9feb6848282963aa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03g3-4109630000-2371d60e702881b24214 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-8729450000-e223ee8d8a68220c25fa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-06vi-8944000000-99c4297b0f97a61c4055 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-066u-6420980000-de8a58be8e7993f3fba3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9532510000-acce9c63da89cfae33d7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9100100000-b6b340ee9a8229e551ec | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03xr-2815090000-1f75f8208fee1665fa0c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-9715010000-75774ec68a0fa55caa15 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03mj-9811000000-7a20e0c1b6fe8912cf8f | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|