| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:26:15 UTC |
|---|
| Update Date | 2020-05-21 16:29:02 UTC |
|---|
| BMDB ID | BMDB0006806 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Propinol adenylate |
|---|
| Description | Propinol adenylate, also known as propinol adenylate, belongs to the class of organic compounds known as 5'-acylphosphoadenosines. These are ribonucleoside derivatives containing an adenoside moiety, where the phosphate group is acylated. Propinol adenylate is possibly soluble (in water) and a strong basic compound (based on its pKa). Propinol adenylate exists in all living organisms, ranging from bacteria to humans. Propinol adenylate participates in a number of enzymatic reactions, within cattle. In particular, Propinol adenylate can be converted into propionic acid; which is catalyzed by the enzyme acyl-CoA synthetase short-chain family member 3, mitochondrial. In addition, Propinol adenylate can be converted into propionic acid; which is mediated by the enzyme acetyl-coenzyme A synthetase 2-like, mitochondrial. In cattle, propinol adenylate is involved in the metabolic pathway called the propanoate metabolism pathway. |
|---|
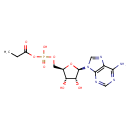
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 5'-Adenylic acid propanoic acid anhydride | ChEBI | | 5'-Adenylic acid propionic acid anhydride | ChEBI | | 5'-O-[Hydroxy(propionyloxy)phosphoryl]adenosine | ChEBI | | Propanoyl-adenosine monophosphate | ChEBI | | Propionyl-adenosine monophosphate | ChEBI | | Propionyl-AMP | ChEBI | | Propionyladenylate | ChEBI | | 5'-Adenylate propanoate anhydride | Generator | | 5'-Adenylate propionate anhydride | Generator | | Propanoyl-adenosine monophosphoric acid | Generator | | Propionyl-adenosine monophosphoric acid | Generator | | Propionyladenylic acid | Generator | | Propinol adenylic acid | Generator |
|
|---|
| Chemical Formula | C13H18N5O8P |
|---|
| Average Molecular Weight | 403.2845 |
|---|
| Monoisotopic Molecular Weight | 403.089299089 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(propanoyloxy)phosphinic acid |
|---|
| Traditional Name | propionyl-AMP |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CCC(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C13H18N5O8P/c1-2-7(19)26-27(22,23)24-3-6-9(20)10(21)13(25-6)18-5-17-8-11(14)15-4-16-12(8)18/h4-6,9-10,13,20-21H,2-3H2,1H3,(H,22,23)(H2,14,15,16)/t6-,9-,10-,13-/m1/s1 |
|---|
| InChI Key | ZGNGGJLVZZHLQM-ZRFIDHNTSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 5'-acylphosphoadenosines. These are ribonucleoside derivatives containing an adenoside moiety, where the phosphate group is acylated. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine ribonucleotides |
|---|
| Direct Parent | 5'-acylphosphoadenosines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 5'-acylphosphoadenosine
- Pentose phosphate
- Pentose-5-phosphate
- N-glycosyl compound
- Glycosyl compound
- 6-aminopurine
- Pentose monosaccharide
- Monosaccharide phosphate
- Imidazopyrimidine
- Purine
- Acyl phosphate
- Monoalkyl phosphate
- Aminopyrimidine
- Imidolactam
- Alkyl phosphate
- Monosaccharide
- Pyrimidine
- N-substituted imidazole
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Imidazole
- Tetrahydrofuran
- Heteroaromatic compound
- Azole
- 1,2-diol
- Amino acid or derivatives
- Carboxylic acid salt
- Secondary alcohol
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Organonitrogen compound
- Hydrocarbon derivative
- Organic oxygen compound
- Organic salt
- Carbonyl group
- Organic oxide
- Organopnictogen compound
- Organic nitrogen compound
- Amine
- Alcohol
- Primary amine
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0lya-5913000000-c9cc25ab5bf7cdb806ab | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-01r2-8923410000-ad6814a72184f59e8542 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-5914100000-91cfd0c5b4d29b3488b9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-3900000000-16aca9edc0f28599b061 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-2900000000-2e68af310b047b1bdb7c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-4904300000-5d204fda744d81dc0866 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-003r-6900000000-dd107e3714dfcd94b540 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-003r-9500000000-0396a7faf046b6610b7a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0309300000-552e044f229a538160dc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-1910000000-3030d909025ec83d805c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-2900000000-25b5fd21f4d139eaa4f5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-4009300000-68e440eecae9e4f48317 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-461bd77745c544eb03d1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9101000000-f61d576d5d36bebef467 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|