| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:27:30 UTC |
|---|
| Update Date | 2020-04-22 15:19:03 UTC |
|---|
| BMDB ID | BMDB0006902 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Cobinamide |
|---|
| Description | Cobinamide belongs to the class of organic compounds known as metallotetrapyrroles. These are polycyclic compounds containing a tetrapyrrole skeleton combined with a metal atom. Based on a literature review a significant number of articles have been published on Cobinamide. |
|---|
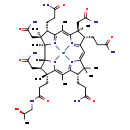
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Cbi | ChEBI | | Cob(I)inamide | ChEBI | | Cobinamid | ChEBI | | Cobinamide dihydrate | HMDB | | Diaquocobinamide | HMDB |
|
|---|
| Chemical Formula | C48H72CoN11O8 |
|---|
| Average Molecular Weight | 990.0874 |
|---|
| Monoisotopic Molecular Weight | 989.489733529 |
|---|
| IUPAC Name | [(1R,2R,3S,4S,6Z,8S,9S,11Z,14S,18R,19R)-4,9,14-tris(2-carbamoylethyl)-3,8,19-tris(carbamoylmethyl)-18-(2-{[(2R)-2-hydroxypropyl]carbamoyl}ethyl)-2,3,6,8,13,13,16,18-octamethyl-20,21,22,23-tetraazapentacyclo[15.2.1.1^{2,5}.1^{7,10}.1^{12,15}]tricosa-5(23),6,10(22),11,15(21),16-hexaen-20-yl]cobaltbis(ylium) |
|---|
| Traditional Name | [(1R,2R,3S,4S,6Z,8S,9S,11Z,14S,18R,19R)-4,9,14-tris(2-carbamoylethyl)-3,8,19-tris(carbamoylmethyl)-18-(2-{[(2R)-2-hydroxypropyl]carbamoyl}ethyl)-2,3,6,8,13,13,16,18-octamethyl-20,21,22,23-tetraazapentacyclo[15.2.1.1^{2,5}.1^{7,10}.1^{12,15}]tricosa-5(23),6,10(22),11,15(21),16-hexaen-20-yl]cobaltbis(ylium) |
|---|
| CAS Registry Number | 13497-85-3 |
|---|
| SMILES | [H][C@@]12[C@H](CC(N)=O)[C@@](C)(CCC(=O)NC[C@@H](C)O)C3=C(C)C4=[N]5C(=CC6=[N]7C(=C(C)C8=[N]([C@]1(C)[C@@](C)(CC(N)=O)[C@@H]8CCC(N)=O)[Co++]57N23)[C@@](C)(CC(N)=O)[C@@H]6CCC(N)=O)C(C)(C)[C@@H]4CCC(N)=O |
|---|
| InChI Identifier | InChI=1S/C48H73N11O8.Co/c1-23(60)22-55-38(67)16-17-45(6)29(18-35(52)64)43-48(9)47(8,21-37(54)66)28(12-15-34(51)63)40(59-48)25(3)42-46(7,20-36(53)65)26(10-13-32(49)61)30(56-42)19-31-44(4,5)27(11-14-33(50)62)39(57-31)24(2)41(45)58-43;/h19,23,26-29,43,60H,10-18,20-22H2,1-9H3,(H14,49,50,51,52,53,54,55,56,57,58,59,61,62,63,64,65,66,67);/q;+3/p-1/t23-,26-,27-,28-,29+,43-,45-,46+,47+,48+;/m1./s1 |
|---|
| InChI Key | FEESAGIUMZGLMF-JFYQDRLCSA-M |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as metallotetrapyrroles. These are polycyclic compounds containing a tetrapyrrole skeleton combined with a metal atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Tetrapyrroles and derivatives |
|---|
| Sub Class | Metallotetrapyrroles |
|---|
| Direct Parent | Metallotetrapyrroles |
|---|
| Alternative Parents | |
|---|
| Substituents | - Metallotetrapyrrole skeleton
- Fatty amide
- Fatty acyl
- Pyrrolidine
- Pyrroline
- Carboxamide group
- Ketimine
- Primary carboxylic acid amide
- Secondary alcohol
- Secondary carboxylic acid amide
- Carbene-type 1,3-dipolar compound
- Azacycle
- Carboxylic acid derivative
- Organic transition metal salt
- Alcohol
- Organic cobalt salt
- Organic oxide
- Organopnictogen compound
- Carbonyl group
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Imine
- Organic nitrogen compound
- Organic salt
- Hydrocarbon derivative
- Organic cation
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|