| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:46:29 UTC |
|---|
| Update Date | 2020-06-04 19:27:15 UTC |
|---|
| BMDB ID | BMDB0007890 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z)) |
|---|
| Description | PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z)), also known as pc(14:0/22:5(4z,7z,10z,13z,16z)) or PC(14:0/22:5), belongs to the class of organic compounds known as phosphatidylcholines. These are glycerophosphocholines in which the two free -OH are attached to one fatty acid each through an ester linkage. Thus, PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z)) is considered to be a glycerophosphocholine lipid molecule. PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z)) is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z)) exists in all eukaryotes, ranging from yeast to humans. PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z)) participates in a number of enzymatic reactions, within cattle. In particular, S-Adenosylhomocysteine and PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z)) can be biosynthesized from S-adenosylmethionine and pe-nme2(14:0/22:5(4Z,7Z,10Z,13Z,16Z)); which is catalyzed by the enzyme phosphatidylethanolamine N-methyltransferase. Furthermore, Cytidine monophosphate and PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z)) can be biosynthesized from CDP-choline and DG(14:0/22:5(4Z,7Z,10Z,13Z,16Z)/0:0) through the action of the enzyme choline/ethanolaminephosphotransferase. Finally, PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z)) and L-serine can be converted into choline and PS(14:0/22:5(4Z,7Z,10Z,13Z,16Z)); which is catalyzed by the enzyme phosphatidylserine synthase. In cattle, PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z)) is involved in a couple of metabolic pathways, which include phosphatidylcholine biosynthesis PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z)) pathway and phosphatidylethanolamine biosynthesis pe(14:0/22:5(4Z,7Z,10Z,13Z,16Z)) pathway. |
|---|
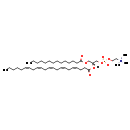
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| GPCho(36:5) | HMDB | | PC(14:0/22:5) | HMDB | | Phosphatidylcholine(36:5) | HMDB | | Lecithin | HMDB | | Phosphatidylcholine(14:0/22:5) | HMDB | | PC(36:5) | HMDB | | 1-Myristoyl-2-osbondoyl-sn-glycero-3-phosphocholine | HMDB | | 1-Tetradecanoyl-2-(4Z,7Z,10Z,13Z,16Z-docosapentaenoyl)-sn-glycero-3-phosphocholine | HMDB | | GPCho(14:0/22:5) | HMDB | | PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z)) | Lipid Annotator |
|
|---|
| Chemical Formula | C44H78NO8P |
|---|
| Average Molecular Weight | 780.0658 |
|---|
| Monoisotopic Molecular Weight | 779.546504989 |
|---|
| IUPAC Name | (2-{[(2R)-2-[(4Z,7Z,10Z,13Z,16Z)-docosa-4,7,10,13,16-pentaenoyloxy]-3-(tetradecanoyloxy)propyl phosphonato]oxy}ethyl)trimethylazanium |
|---|
| Traditional Name | lecithin |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CCCCCCCCCCCCCC(=O)OC[C@]([H])(COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CC\C=C/C\C=C/C\C=C/C\C=C/C\C=C/CCCCC |
|---|
| InChI Identifier | InChI=1S/C44H78NO8P/c1-6-8-10-12-14-16-18-19-20-21-22-23-24-25-27-29-31-33-35-37-44(47)53-42(41-52-54(48,49)51-39-38-45(3,4)5)40-50-43(46)36-34-32-30-28-26-17-15-13-11-9-7-2/h14,16,19-20,22-23,25,27,31,33,42H,6-13,15,17-18,21,24,26,28-30,32,34-41H2,1-5H3/b16-14-,20-19-,23-22-,27-25-,33-31-/t42-/m1/s1 |
|---|
| InChI Key | MGUTYWPZCZBAGE-JZDIRDTQSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phosphatidylcholines. These are glycerophosphocholines in which the two free -OH are attached to one fatty acid each through an ester linkage. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphocholines |
|---|
| Direct Parent | Phosphatidylcholines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Diacylglycero-3-phosphocholine
- Phosphocholine
- Fatty acid ester
- Dialkyl phosphate
- Dicarboxylic acid or derivatives
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Fatty acyl
- Quaternary ammonium salt
- Tetraalkylammonium salt
- Carboxylic acid ester
- Carboxylic acid derivative
- Organic oxygen compound
- Organic nitrogen compound
- Carbonyl group
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Amine
- Organic salt
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected and Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Intracellular membrane
- Membrane
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03ei-9154331300-fe605cf40fb850b6d995 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-5594120100-73a2f90b2c9093e8c79c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0019-8292010100-499249892f080d880c49 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0093000300-6bf32e98a7c91ea9768d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-0192001000-85d51b96d6c99e5af921 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-7297000000-4bf9b9fa3f173a9df0b7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0000000900-11abb7219d74a5a60a04 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0600000900-8b887b2cddb30aa5f84a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ff0-1900230300-06c6720b1d231f99f823 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0000000090-be732a4aa7f691f68131 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0000000190-37d5d9b145362a874a18 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014j-0900449110-a86a54619f09a4ddbc6e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0000000090-d1fecbe733b4fab2e6fb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-0011000090-63cbf52d035bcf4b7043 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01t9-0099000090-e7a83072a8b00fcc7e8c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0000000900-6ac0ce44c5d362b404ab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-0034030900-3548cc7c5c8f42159295 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-4392000000-69908c9669a9069d6308 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000000900-82395bc23558c5e73383 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0000000900-d73ebce581a3424e67e3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0zi1-0202649400-7477ed35d87f5516786a | View in MoNA |
|---|
|
|---|