| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-10-03 18:14:19 UTC |
|---|
| Update Date | 2020-04-22 15:43:10 UTC |
|---|
| BMDB ID | BMDB0011188 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
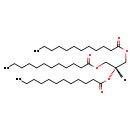
| Common Name | TG(12:0/12:0/12:0) |
|---|
| Description | TG(12:0/12:0/12:0), also known as tg(12:0/12:0/12:0) or tg(12:0/12:0/12:0), belongs to the class of organic compounds known as tg(12:0/12:0/12:0)s. These are glycerides consisting of three fatty acid chains covalently bonded to a glycerol molecule through ester linkages. Thus, TG(12:0/12:0/12:0) is considered to be a triradylglycerol lipid molecule. TG(12:0/12:0/12:0) is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. TG(12:0/12:0/12:0) exists in all eukaryotes, ranging from yeast to humans. TG(12:0/12:0/12:0) can be biosynthesized from DG(12:0/12:0/0:0) and lauroyl-CoA through its interaction with the enzyme diacylglycerol O-acyltransferase. In cattle, TG(12:0/12:0/12:0) is involved in the metabolic pathway called de novo tg(12:0/12:0/12:0) biosynthesis TG(12:0/12:0/12:0) pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,2,3-Tridodecanoylglycerol | HMDB | | 1,2,3-Trilauroylglycerol | HMDB | | Dodecanoic acid 1,2,3-propanetriyl ester | HMDB | | Glycerin trilaurate | HMDB | | Glycerol trilaurate | HMDB | | Glyceryl tridodecanoate | HMDB | | Glyceryl trilaurate | HMDB | | Lauric acid triglyceride | HMDB | | Lauric acid triglycerin ester | HMDB | | Propane-1,2,3-triyl trilaurate | HMDB | | TG 12:0/12:0/12:0 | HMDB | | Tridodecanoin | HMDB | | Tridodecanoyl glycerol | HMDB | | Tridodecanoylglycerol | HMDB | | Trilauroylglycerol | HMDB | | Dodecanoate 1,2,3-propanetriyl ester | HMDB | | Glycerin trilauric acid | HMDB | | Glycerol trilauric acid | HMDB | | Glyceryl tridodecanoic acid | HMDB | | Glyceryl trilauric acid | HMDB | | Laate triglyceride | HMDB | | Laic acid triglyceride | HMDB | | Laate triglycerin ester | HMDB | | Laic acid triglycerin ester | HMDB | | Propane-1,2,3-triyl trilauric acid | HMDB | | Glycerin trilaate | HMDB | | Glycerin trilaic acid | HMDB | | Glycerol trilaate | HMDB | | Glycerol trilaic acid | HMDB | | Glyceryl trilaate | HMDB | | Glyceryl trilaic acid | HMDB | | Propane-1,2,3-triyl trilaate | HMDB | | Propane-1,2,3-triyl trilaic acid | HMDB | | Tracylglycerol(12:0/12:0/12:0) | HMDB | | Tracylglycerol(36:0) | HMDB | | 1-Dodecanoyl-2-dodecanoyl-3-dodecanoyl-glycerol | HMDB | | Triglyceride | HMDB | | TG(36:0) | HMDB | | TAG(36:0) | HMDB | | TAG(12:0/12:0/12:0) | HMDB | | Triacylglycerol | HMDB | | Trilaurin | HMDB | | TG(12:0/12:0/12:0) | Lipid Annotator, ChEBI |
|
|---|
| Chemical Formula | C39H74O6 |
|---|
| Average Molecular Weight | 639.0013 |
|---|
| Monoisotopic Molecular Weight | 638.5485401 |
|---|
| IUPAC Name | 1,3-bis(dodecanoyloxy)propan-2-yl dodecanoate |
|---|
| Traditional Name | 1,3-bis(dodecanoyloxy)propan-2-yl dodecanoate |
|---|
| CAS Registry Number | 538-24-9 |
|---|
| SMILES | [H]C(COC(=O)CCCCCCCCCCC)(COC(=O)CCCCCCCCCCC)OC(=O)CCCCCCCCCCC |
|---|
| InChI Identifier | InChI=1S/C39H74O6/c1-4-7-10-13-16-19-22-25-28-31-37(40)43-34-36(45-39(42)33-30-27-24-21-18-15-12-9-6-3)35-44-38(41)32-29-26-23-20-17-14-11-8-5-2/h36H,4-35H2,1-3H3 |
|---|
| InChI Key | VMPHSYLJUKZBJJ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triacylglycerols. These are glycerides consisting of three fatty acid chains covalently bonded to a glycerol molecule through ester linkages. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Triradylcglycerols |
|---|
| Direct Parent | Triacylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triacyl-sn-glycerol
- Tricarboxylic acid or derivatives
- Fatty acid ester
- Fatty acyl
- Carboxylic acid ester
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Adiposome
- Cell membrane
- Membrane
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-001l-3751900000-186ab5f4aeb9dcd0e97b | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-001l-3751900000-186ab5f4aeb9dcd0e97b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0000009000-f0a58fa5fad480d99baf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-0000009000-f0a58fa5fad480d99baf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0000907000-ab658a63ebc1bf1cb2e9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0000009000-5dfb05e6553aa8729621 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-0000009000-5dfb05e6553aa8729621 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4q-0090909000-7e0e947d313b7e4974cb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0000009000-65bd852687bb227267a1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-0000009000-65bd852687bb227267a1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0100907000-9ebd50ecc56178ec88c4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0330709000-f808d6e66ac99e938d36 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004s-0390301000-f6dbb62ac5df927a6e8d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0092-1950300000-c6be0d26c1f534f7d613 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000009000-b04629da917188e944f0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0000009000-b04629da917188e944f0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-0000009000-b04629da917188e944f0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-1110329000-7fa56409688ae35da8c0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-4513391000-709b5c1ed2eb8fcfcf51 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0540-1790210000-41ed924482264ba34297 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 90 MHz, CDCl3, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 25.16 MHz, CDCl3, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|