| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-10-03 18:35:01 UTC |
|---|
| Update Date | 2020-05-05 18:38:33 UTC |
|---|
| BMDB ID | BMDB0012253 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Maltohexaose |
|---|
| Description | Maltohexaose belongs to the class of organic compounds known as oligosaccharides. These are carbohydrates made up of 3 to 10 monosaccharide units linked to each other through glycosidic bonds. Based on a literature review very few articles have been published on Maltohexaose. |
|---|
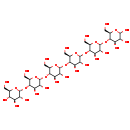
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| alpha-D-GLCP-(1->4)-alpha-D-GLCP-(1->4)-alpha-D-GLCP-(1->4)-alpha-D-GLCP-(1->4)-alpha-D-GLCP-(1->4)-alpha-D-GLCP | ChEBI | | WURCS=2.0/1,6,5/[a2122h-1a_1-5]/1-1-1-1-1-1/a4-b1_b4-c1_c4-D1_d4-e1_e4-F1 | ChEBI | | a-D-GLCP-(1->4)-a-D-GLCP-(1->4)-a-D-GLCP-(1->4)-a-D-GLCP-(1->4)-a-D-GLCP-(1->4)-a-D-GLCP | Generator | | Α-D-GLCP-(1->4)-α-D-GLCP-(1->4)-α-D-GLCP-(1->4)-α-D-GLCP-(1->4)-α-D-GLCP-(1->4)-α-D-GLCP | Generator | | Maltohexanose DP6 | HMDB | | a-Maltohexaose | HMDB | | Α-maltohexaose | HMDB | | Maltohexaose | MeSH |
|
|---|
| Chemical Formula | C36H62O31 |
|---|
| Average Molecular Weight | 990.8589 |
|---|
| Monoisotopic Molecular Weight | 990.327505266 |
|---|
| IUPAC Name | (2R,3R,4S,5S,6R)-2-{[(2R,3S,4R,5R,6R)-6-{[(2R,3S,4R,5R,6R)-6-{[(2R,3S,4R,5R,6R)-6-{[(2R,3S,4R,5R,6R)-4,5-dihydroxy-2-(hydroxymethyl)-6-{[(2R,3S,4R,5R,6S)-4,5,6-trihydroxy-2-(hydroxymethyl)oxan-3-yl]oxy}oxan-3-yl]oxy}-4,5-dihydroxy-2-(hydroxymethyl)oxan-3-yl]oxy}-4,5-dihydroxy-2-(hydroxymethyl)oxan-3-yl]oxy}-4,5-dihydroxy-2-(hydroxymethyl)oxan-3-yl]oxy}-6-(hydroxymethyl)oxane-3,4,5-triol |
|---|
| Traditional Name | α-maltohexaose |
|---|
| CAS Registry Number | 34620-77-4 |
|---|
| SMILES | OC[C@H]1O[C@H](O[C@H]2[C@H](O)[C@@H](O)[C@@H](O[C@H]3[C@H](O)[C@@H](O)[C@@H](O[C@H]4[C@H](O)[C@@H](O)[C@@H](O[C@H]5[C@H](O)[C@@H](O)[C@@H](O[C@H]6[C@H](O)[C@@H](O)[C@@H](O)O[C@@H]6CO)O[C@@H]5CO)O[C@@H]4CO)O[C@@H]3CO)O[C@@H]2CO)[C@H](O)[C@@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C36H62O31/c37-1-7-13(43)14(44)21(51)32(58-7)64-27-9(3-39)60-34(23(53)16(27)46)66-29-11(5-41)62-36(25(55)18(29)48)67-30-12(6-42)61-35(24(54)19(30)49)65-28-10(4-40)59-33(22(52)17(28)47)63-26-8(2-38)57-31(56)20(50)15(26)45/h7-56H,1-6H2/t7-,8-,9-,10-,11-,12-,13-,14+,15-,16-,17-,18-,19-,20-,21-,22-,23-,24-,25-,26-,27-,28-,29-,30-,31+,32-,33-,34-,35-,36-/m1/s1 |
|---|
| InChI Key | OCIBBXPLUVYKCH-QXVNYKTNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as oligosaccharides. These are carbohydrates made up of 3 to 10 monosaccharide units linked to each other through glycosidic bonds. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Oligosaccharides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Oligosaccharide
- O-glycosyl compound
- Glycosyl compound
- Oxane
- Secondary alcohol
- Hemiacetal
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Acetal
- Hydrocarbon derivative
- Primary alcohol
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-022j-0605538059-ecfce7e80c18b4fda045 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-08gv-0807759053-231ffd53e9bc79e901d3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08gu-0906223021-9eb02fa61e039b7ce67e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00dr-0211212129-cd7ad365fd2b2cb0df46 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-0412213029-0e4d854ce072c0cff03c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004l-1926122112-e152a4323e2e58d783c9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000000019-c3268d693555664f49d9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a70-7213001069-cf99e3064fb8b32232ae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a6r-9733155373-ac507562bffac29d8754 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-006x-0200001019-3ee7406b8db4382b4060 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0234-0402101039-54da292aa05f0f5e7d22 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0007-9601000005-6d507b2b423ff6722c6f | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|