| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-10-03 18:35:29 UTC |
|---|
| Update Date | 2020-04-22 15:49:27 UTC |
|---|
| BMDB ID | BMDB0012281 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Portulacaxanthin II |
|---|
| Description | Portulacaxanthin II belongs to the class of organic compounds known as tyrosine and derivatives. Tyrosine and derivatives are compounds containing tyrosine or a derivative thereof resulting from reaction of tyrosine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. Based on a literature review very few articles have been published on Portulacaxanthin II. |
|---|
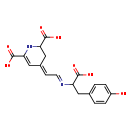
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Tyrosine-betaxanthin | HMDB | | (4E)-4-[(2E)-2-{[1-carboxy-2-(4-hydroxyphenyl)ethyl]imino}ethylidene]-1,2,3,4-tetrahydropyridine-2,6-dicarboxylate | Generator |
|
|---|
| Chemical Formula | C18H18N2O7 |
|---|
| Average Molecular Weight | 374.3447 |
|---|
| Monoisotopic Molecular Weight | 374.11140094 |
|---|
| IUPAC Name | (4E)-4-[(2E)-2-{[1-carboxy-2-(4-hydroxyphenyl)ethyl]imino}ethylidene]-1,2,3,4-tetrahydropyridine-2,6-dicarboxylic acid |
|---|
| Traditional Name | (4E)-4-[(2E)-2-{[1-carboxy-2-(4-hydroxyphenyl)ethyl]imino}ethylidene]-2,3-dihydro-1H-pyridine-2,6-dicarboxylic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | OC(=O)C(CC1=CC=C(O)C=C1)\N=C\C=C1/CC(NC(=C1)C(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C18H18N2O7/c21-12-3-1-10(2-4-12)7-13(16(22)23)19-6-5-11-8-14(17(24)25)20-15(9-11)18(26)27/h1-6,8,13,15,20-21H,7,9H2,(H,22,23)(H,24,25)(H,26,27)/b11-5-,19-6+ |
|---|
| InChI Key | MBFJCQLVRQZZOV-KRDOMJLVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as tyrosine and derivatives. Tyrosine and derivatives are compounds containing tyrosine or a derivative thereof resulting from reaction of tyrosine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Tyrosine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Tyrosine or derivatives
- Phenylalanine or derivatives
- 3-phenylpropanoic-acid
- Alpha-amino acid
- Amphetamine or derivatives
- Tricarboxylic acid or derivatives
- 1-hydroxy-2-unsubstituted benzenoid
- Phenol
- Tetrahydropyridine
- Monocyclic benzene moiety
- Hydropyridine
- Benzenoid
- Amino acid
- Shiff base
- Aldimine
- Carboxylic acid
- Secondary aliphatic amine
- Enamine
- Azacycle
- Secondary amine
- Organoheterocyclic compound
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Organic oxide
- Organic oxygen compound
- Imine
- Organonitrogen compound
- Organooxygen compound
- Hydrocarbon derivative
- Organopnictogen compound
- Carbonyl group
- Organic nitrogen compound
- Amine
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-2935000000-63da66d3c21c5ad9bee6 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-0002-7622189000-c172f7a7c11e7495a482 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-056r-0209000000-fc4975e1e299f159766b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0kc6-0924000000-889c66a4b1c0d13090e7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-3900000000-96b8325f01750e55a3c5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0019000000-f17fcecdea5740aa6a01 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0c30-0369000000-dd49d12eb56ce1705556 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00si-5910000000-cfc203bb338c72db28e2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0009000000-86324c4b89f5c5f942c9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01u0-0479000000-e868d4ee5a1d67f5fbb4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0mjj-1941000000-8ea5a0b1178896c8d1a3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00c0-0039000000-ecc3d799b126db513783 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00c0-0159000000-c5f05401670636b6f2fb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-006x-7391000000-d93e54266a51bb0129d0 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|