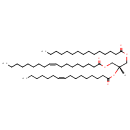| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2018-08-29 17:19:34 UTC |
|---|
| Update Date | 2020-06-04 20:23:39 UTC |
|---|
| BMDB ID | BMDB0063293 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | TG(15:0/16:1(9Z)/18:1(9Z))[iso6] |
|---|
| Description | TG(15:0/16:1(9Z)/18:1(9Z))[iso6] belongs to the class of organic compounds known as triacylglycerols. These are glycerides consisting of three fatty acid chains covalently bonded to a glycerol molecule through ester linkages. Thus, TG(15:0/16:1(9Z)/18:1(9Z))[iso6] is considered to be a triradylglycerol lipid molecule. TG(15:0/16:1(9Z)/18:1(9Z))[iso6] is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Pentadecanoyl-2-(9Z-hexadecenoyl)-3-(9Z-octadecenoyl)-glycerol | HMDB | | 1-Pentadecanoyl-2-palmitoleoyl-3-oleoyl-glycerol | HMDB | | TAG(15:0/16:1/18:1) | HMDB | | TAG(49:2) | HMDB | | TG(15:0/16:1/18:1) | HMDB | | TG(49:2) | HMDB | | Tracylglycerol(15:0/16:1/18:1) | HMDB | | Tracylglycerol(49:2) | HMDB | | Triacylglycerol | HMDB | | Triglyceride | HMDB | | TG(15:0/16:1(9Z)/18:1(9Z)) | Lipid Annotator |
|
|---|
| Chemical Formula | C52H96O6 |
|---|
| Average Molecular Weight | 817.334 |
|---|
| Monoisotopic Molecular Weight | 816.720690811 |
|---|
| IUPAC Name | (2S)-2-[(9Z)-hexadec-9-enoyloxy]-3-(pentadecanoyloxy)propyl (9Z)-octadec-9-enoate |
|---|
| Traditional Name | (2S)-2-[(9Z)-hexadec-9-enoyloxy]-3-(pentadecanoyloxy)propyl (9Z)-octadec-9-enoate |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | [H][C@](COC(=O)CCCCCCCCCCCCCC)(COC(=O)CCCCCCC\C=C/CCCCCCCC)OC(=O)CCCCCCC\C=C/CCCCCC |
|---|
| InChI Identifier | InChI=1S/C52H96O6/c1-4-7-10-13-16-19-22-25-26-28-30-33-36-39-42-45-51(54)57-48-49(47-56-50(53)44-41-38-35-32-29-24-21-18-15-12-9-6-3)58-52(55)46-43-40-37-34-31-27-23-20-17-14-11-8-5-2/h20,23,25-26,49H,4-19,21-22,24,27-48H2,1-3H3/b23-20-,26-25-/t49-/m0/s1 |
|---|
| InChI Key | ZSHVDBVWHRXKEV-OPRAGXQSSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triacylglycerols. These are glycerides consisting of three fatty acid chains covalently bonded to a glycerol molecule through ester linkages. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Triradylcglycerols |
|---|
| Direct Parent | Triacylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triacyl-sn-glycerol
- Tricarboxylic acid or derivatives
- Fatty acid ester
- Fatty acyl
- Carboxylic acid ester
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Detected and Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Adiposome
- Cell membrane
- Membrane
|
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0000000090-65d89e30ece759308597 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0000000090-65d89e30ece759308597 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-042r-0000090020-8d98aaeed35a34988d55 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0iru-0090010010-eaba81da9b56382bff02 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01x3-0090000000-e57c4e731dfa5a4ba15e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000x-1090000000-ec31ecb75b9a9ea5767e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0000000090-39543117b1776265cab9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-0000000090-39543117b1776265cab9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01k0-0040090040-5cc1278899ad2632be67 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0000000090-3b49e6675437b4cdaec8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0000000090-3b49e6675437b4cdaec8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-02e0-0010090020-caa139b42b109c6287fe | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0170-5260082490-74c51a968921e7272f79 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-052r-9220021300-7d881ef8d56ecf95cdd0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0fdx-6694031200-9b792005d970161b616a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0090060060-9368460a2310940ba8cd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-003r-0091010000-b14426cf9456de2227dd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001l-1091000000-ee7ffe8bc98d8362718d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000000090-1cd4c0c80270bcd9ea08 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0000000090-1cd4c0c80270bcd9ea08 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0000000090-1cd4c0c80270bcd9ea08 | View in MoNA |
|---|
|
|---|