| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-03 19:41:40 UTC |
|---|
| Update Date | 2020-04-22 15:57:11 UTC |
|---|
| BMDB ID | BMDB0064115 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Tyrosyl-Glutamine |
|---|
| Description | Tyrosyl-Glutamine, also known as y-Q dipeptide or tyr-GLN, belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. Based on a literature review very few articles have been published on Tyrosyl-Glutamine. |
|---|
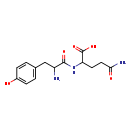
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| L-Tyrosyl-L-glutamine | HMDB | | Tyr-GLN | HMDB | | Tyrosine glutamine dipeptide | HMDB | | Tyrosine-glutamine dipeptide | HMDB | | Tyrosylglutamine | HMDB | | Y-Q dipeptide | HMDB | | YQ dipeptide | HMDB | | 2-{[2-amino-1-hydroxy-3-(4-hydroxyphenyl)propylidene]amino}-4-(C-hydroxycarbonimidoyl)butanoate | HMDB |
|
|---|
| Chemical Formula | C14H19N3O5 |
|---|
| Average Molecular Weight | 309.3178 |
|---|
| Monoisotopic Molecular Weight | 309.132470733 |
|---|
| IUPAC Name | 2-[2-amino-3-(4-hydroxyphenyl)propanamido]-4-carbamoylbutanoic acid |
|---|
| Traditional Name | 2-[2-amino-3-(4-hydroxyphenyl)propanamido]-4-carbamoylbutanoic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | NC(CC1=CC=C(O)C=C1)C(=O)NC(CCC(N)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C14H19N3O5/c15-10(7-8-1-3-9(18)4-2-8)13(20)17-11(14(21)22)5-6-12(16)19/h1-4,10-11,18H,5-7,15H2,(H2,16,19)(H,17,20)(H,21,22) |
|---|
| InChI Key | UBAQSAUDKMIEQZ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Dipeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-dipeptide
- Tyrosine or derivatives
- Phenylalanine or derivatives
- Glutamine or derivatives
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- Alpha-amino acid amide
- Alpha-amino acid or derivatives
- Amphetamine or derivatives
- 1-hydroxy-2-unsubstituted benzenoid
- Aralkylamine
- Phenol
- Monocyclic benzene moiety
- Fatty amide
- Fatty acyl
- Benzenoid
- Amino acid or derivatives
- Carboxamide group
- Amino acid
- Secondary carboxylic acid amide
- Primary carboxylic acid amide
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Hydrocarbon derivative
- Primary aliphatic amine
- Amine
- Organic oxygen compound
- Carbonyl group
- Organic oxide
- Organic nitrogen compound
- Organonitrogen compound
- Organooxygen compound
- Organopnictogen compound
- Primary amine
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01wu-4940000000-08b5f699e1f8a999fb93 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-000i-2905000000-f9506782226a7b6d82a0 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01ox-0592000000-dd211f04fdc3f5ef353f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kr-0930000000-8e8ad194206afb625ae4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-2900000000-59f910b4f094fa644c93 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0169000000-66700e0077326e0f5ee4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01oy-3982000000-279fd254223d676207b2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9600000000-687e806511fa7b2bfdf8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a6u-0956000000-57445ce689cc9bec14a1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01r6-3910000000-5b9ecc8c8eb4602d3322 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-006x-9800000000-74b857c8465fcbab91a2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0937000000-d3882b0ce3346a338d14 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-052r-0910000000-71f7be548f022cb19536 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-5900000000-66549c4bb2cb7c0ccca5 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|