| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-03 19:44:13 UTC |
|---|
| Update Date | 2020-04-22 15:57:26 UTC |
|---|
| BMDB ID | BMDB0064158 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | gamma-Glutamylproline |
|---|
| Description | gamma-Glutamylproline, also known as ge-p dipeptide or gglu-pro, belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. Based on a literature review very few articles have been published on gamma-Glutamylproline. |
|---|
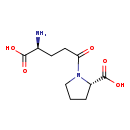
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| g-Glutamylproline | Generator | | Γ-glutamylproline | Generator | | gamma-Glutamate proline dipeptide | HMDB | | gamma-Glutamate-proline dipeptide | HMDB | | GE-p dipeptide | HMDB | | GEP dipeptide | HMDB | | GGlu-pro | HMDB | | L-gamma-Glutamyl-L-proline | HMDB | | Γ-glu-pro | HMDB | | Γ-L-glu-L-pro | HMDB | | Γ-L-glutamyl-L-proline | HMDB | | L-Γ-glutamyl-L-proline | HMDB | | gamma-Glu-pro | HMDB | | gamma-L-Glu-L-pro | HMDB | | gamma-L-Glutamyl-L-proline | HMDB | | (2S)-1-[(4S)-4-Amino-4-carboxybutanoyl]pyrrolidine-2-carboxylate | HMDB | | gamma-Glutamylproline | HMDB |
|
|---|
| Chemical Formula | C10H16N2O5 |
|---|
| Average Molecular Weight | 244.247 |
|---|
| Monoisotopic Molecular Weight | 244.105921623 |
|---|
| IUPAC Name | (2S)-1-[(4S)-4-amino-4-carboxybutanoyl]pyrrolidine-2-carboxylic acid |
|---|
| Traditional Name | (2S)-1-[(4S)-4-amino-4-carboxybutanoyl]pyrrolidine-2-carboxylic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | N[C@@H](CCC(=O)N1CCC[C@H]1C(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C10H16N2O5/c11-6(9(14)15)3-4-8(13)12-5-1-2-7(12)10(16)17/h6-7H,1-5,11H2,(H,14,15)(H,16,17)/t6-,7-/m0/s1 |
|---|
| InChI Key | VBCZKAGVUKCANO-BQBZGAKWSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Dipeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-dipeptide
- Gamma-glutamyl alpha-amino acid
- Glutamine or derivatives
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- N-acyl-l-alpha-amino acid
- Proline or derivatives
- Alpha-amino acid or derivatives
- L-alpha-amino acid
- Alpha-amino acid
- N-acylpyrrolidine
- Pyrrolidine carboxylic acid
- Pyrrolidine carboxylic acid or derivatives
- Heterocyclic fatty acid
- Dicarboxylic acid or derivatives
- Fatty acyl
- Fatty acid
- Tertiary carboxylic acid amide
- Pyrrolidine
- Amino acid
- Amino acid or derivatives
- Carboxamide group
- Organoheterocyclic compound
- Carboxylic acid
- Azacycle
- Amine
- Primary aliphatic amine
- Organic nitrogen compound
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Organonitrogen compound
- Organooxygen compound
- Organic oxygen compound
- Primary amine
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002b-0980000000-8a1cd401fdb5e05dd9c8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000t-2910000000-b827663230992302012d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-066r-9400000000-fbe9d16e1bcb25bc2314 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0007-0690000000-4f6df51ff93ddee6f5c7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0032-2930000000-e0c1993d7d7ea2720576 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-02ml-9400000000-20d089c4dc42b36eee9e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01t9-0790000000-b918b6e73b394134ef36 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-2910000000-b6e1f2141ad7e0653126 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03dm-9500000000-3f3c6b45a991e5971112 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kb-4980000000-a28aaafe8c838eb13225 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00sj-9800000000-07e1a802fb693b887835 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05gi-9200000000-cb00f3642cff0cc1c1c5 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|