| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-04 22:01:05 UTC |
|---|
| Update Date | 2020-05-21 16:29:12 UTC |
|---|
| BMDB ID | BMDB0071029 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
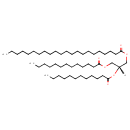
| Common Name | TG(22:0/12:0/13:0) |
|---|
| Description | TG(22:0/12:0/13:0) belongs to the family of triradyglycerols, which are glycerolipids lipids containing a common glycerol backbone to which at least one fatty acyl group is esterified. Their general formula is [R1]OCC(CO[R2])O[R3]. TG(22:0/12:0/13:0) is made up of one docosanoyl(R1), one dodecanoyl(R2), and one tridecanoyl(R3). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-behenoyl-2-lauroyl-3-tridecyloyl-glycerol | SMPDB, HMDB | | TG(47:0) | SMPDB, HMDB | | Tag(22:0/12:0/13:0) | SMPDB, HMDB | | Tag(47:0) | SMPDB, HMDB | | Triacylglycerol(22:0/12:0/13:0) | SMPDB, HMDB | | Triacylglycerol(47:0) | SMPDB, HMDB | | Triacylglycerol | SMPDB, HMDB | | Triglyceride | SMPDB, HMDB | | TG(22:0/12:0/13:0) | SMPDB | | 1-docosanoyl-2-dodecanoyl-3-animal fats-glycerol | Lipid Annotator, HMDB | | Triacylglycerol | Lipid Annotator | | Tracylglycerol(22:0/12:0/13:0) | Lipid Annotator, HMDB | | 1-behenoyl-2-dodecanoyl-3-tridecyloyl-glycerol | Lipid Annotator, HMDB | | TG(47:0) | Lipid Annotator | | TAG(22:0/12:0/13:0) | Lipid Annotator | | Tracylglycerol(47:0) | Lipid Annotator, HMDB | | TAG(47:0) | Lipid Annotator |
|
|---|
| Chemical Formula | C50H96O6 |
|---|
| Average Molecular Weight | 793.312 |
|---|
| Monoisotopic Molecular Weight | 792.720690811 |
|---|
| IUPAC Name | (2R)-2-(dodecanoyloxy)-3-(tridecanoyloxy)propyl docosanoate |
|---|
| Traditional Name | (2R)-2-(dodecanoyloxy)-3-(tridecanoyloxy)propyl docosanoate |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | [H][C@@](COC(=O)CCCCCCCCCCCC)(COC(=O)CCCCCCCCCCCCCCCCCCCCC)OC(=O)CCCCCCCCCCC |
|---|
| InChI Identifier | InChI=1S/C50H96O6/c1-4-7-10-13-16-19-21-22-23-24-25-26-27-28-29-32-34-37-40-43-49(52)55-46-47(56-50(53)44-41-38-35-30-18-15-12-9-6-3)45-54-48(51)42-39-36-33-31-20-17-14-11-8-5-2/h47H,4-46H2,1-3H3/t47-/m1/s1 |
|---|
| InChI Key | SCELHSGUCYEQRX-QZNUWAOFSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triacylglycerols. These are glycerides consisting of three fatty acid chains covalently bonded to a glycerol molecule through ester linkages. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Triradylcglycerols |
|---|
| Direct Parent | Triacylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triacyl-sn-glycerol
- Tricarboxylic acid or derivatives
- Fatty acid ester
- Fatty acyl
- Carboxylic acid ester
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000000090-e6c975ab78e795fcbcf3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0000000090-e6c975ab78e795fcbcf3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f96-0000490300-6bc1037c0b69fbfabb67 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01vw-0859021200-82869ca79a9d68d6d6b1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03kj-0589000000-df85dc15d78c0a963ab7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01p2-4955000000-79d98cbd0cb10159551f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0000000900-7135ed273ab6a5df200f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-0000000900-7135ed273ab6a5df200f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-0004090400-b55f2067cc38a57a6ce3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000000090-88f56269f59b652c7112 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0000000090-88f56269f59b652c7112 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f96-0100490300-c050edcfff1a900dec8e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0000000090-3618b927ca280008090b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0000000090-3618b927ca280008090b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-0000000090-3618b927ca280008090b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-2400010900-7589e5cf33ab587f7667 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00gi-9600021500-bb43073370fb4f780c0c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053u-5954102000-eb9fd61d2717754b829e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0143141900-c226f7fa8aa8c7d10cee | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-0293010000-436b277fabc607ccd9d9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-06tr-1695010000-91e350a4102579c173ad | View in MoNA |
|---|
|
|---|