| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-04 23:38:25 UTC |
|---|
| Update Date | 2020-04-22 16:50:01 UTC |
|---|
| BMDB ID | BMDB0072460 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | TG(22:0/a-15:0/13:0)[rac] |
|---|
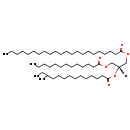
| Description | TG(22:0/a-15:0/13:0) belongs to the family of triradyglycerols, which are glycerolipids lipids containing a common glycerol backbone to which at least one fatty acyl group is esterified. Their general formula is [R1]OCC(CO[R2])O[R3]. TG(22:0/a-15:0/13:0) is made up of one docosanoyl(R1), one 12-methyltetradecanoyl(R2), and one tridecanoyl(R3). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-behenoyl-2-anteisopentadecanoyl-3-tridecyloyl-glycerol | SMPDB, HMDB | | TG(22:0/a-15:0/13:0) | SMPDB, HMDB | | TG(50:0) | SMPDB, HMDB | | Tag(22:0/a-15:0/13:0) | SMPDB, HMDB | | Tag(50:0) | SMPDB, HMDB | | Triacylglycerol(22:0/a-15:0/13:0) | SMPDB, HMDB | | Triacylglycerol(50:0) | SMPDB, HMDB | | Triacylglycerol | SMPDB, HMDB | | Triglyceride | SMPDB, HMDB | | 1-docosanoyl-2-anteisopentadecanoyl-3-animal fats-glycerol | Lipid Annotator, HMDB | | TG(22:0/a-15:0/13:0)[rac] | Lipid Annotator | | Tracylglycerol(50:0) | Lipid Annotator, HMDB | | Tracylglycerol(22:0/a-15:0/13:0) | Lipid Annotator, HMDB |
|
|---|
| Chemical Formula | C53H102O6 |
|---|
| Average Molecular Weight | 835.393 |
|---|
| Monoisotopic Molecular Weight | 834.767641004 |
|---|
| IUPAC Name | (2R)-2-[(12-methyltetradecanoyl)oxy]-3-(tridecanoyloxy)propyl docosanoate |
|---|
| Traditional Name | (2R)-2-[(12-methyltetradecanoyl)oxy]-3-(tridecanoyloxy)propyl docosanoate |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | [H][C@@](COC(=O)CCCCCCCCCCCC)(COC(=O)CCCCCCCCCCCCCCCCCCCCC)OC(=O)CCCCCCCCCCC(C)CC |
|---|
| InChI Identifier | InChI=1S/C53H102O6/c1-5-8-10-12-14-16-18-19-20-21-22-23-24-25-26-28-33-37-41-45-52(55)58-48-50(47-57-51(54)44-40-36-32-27-17-15-13-11-9-6-2)59-53(56)46-42-38-34-30-29-31-35-39-43-49(4)7-3/h49-50H,5-48H2,1-4H3/t49?,50-/m1/s1 |
|---|
| InChI Key | RXRTZAVYOBFTSS-RXFWMZJNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as triacylglycerols. These are glycerides consisting of three fatty acid chains covalently bonded to a glycerol molecule through ester linkages. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Triradylcglycerols |
|---|
| Direct Parent | Triacylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Triacyl-sn-glycerol
- Tricarboxylic acid or derivatives
- Fatty acid ester
- Fatty acyl
- Carboxylic acid ester
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0000000090-1588421d9f01dd529931 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0000000090-1588421d9f01dd529931 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00dz-0000999070-2735192371c64ad1bbb8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0000000090-388332d7b7282853ecb1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0000000090-388332d7b7282853ecb1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00dw-0111999070-6c5b91a950a7700b0b9e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0000000090-90be60e04864d07f44ce | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-0000000090-90be60e04864d07f44ce | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-002e-0009099090-f7d3ede5bbabfd1258f9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-5240111390-245bcc5b96dde4a5f671 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0adj-9120000310-77fe097fb202db64165f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056r-7494001100-3ef01276e8df881c8f81 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0000000090-223de51541149b3b0ccd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-0000000090-223de51541149b3b0ccd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-0000000090-223de51541149b3b0ccd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0053113090-85b460d537bea4a7b74b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01b9-0092000000-fe26a45afd058d09921e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-07il-1294000000-b99a4f0caab925efe901 | View in MoNA |
|---|
|
|---|