| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-10 16:59:47 UTC |
|---|
| Update Date | 2020-04-22 18:55:31 UTC |
|---|
| BMDB ID | BMDB0096014 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
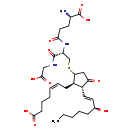
| Common Name | S-(PGJ2)-glutathione |
|---|
| Description | S-(PGJ2)-glutathione belongs to the class of organic compounds known as oligopeptides. These are organic compounds containing a sequence of between three and ten alpha-amino acids joined by peptide bonds. Based on a literature review a significant number of articles have been published on S-(PGJ2)-glutathione. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 9-(S-Glutathionyl)-prostaglandin J(,2) | HMDB | | GSPGJ2 | HMDB | | (5Z)-7-[(1R,2R)-5-{[(2S)-2-{[(4R)-4-amino-4-carboxy-1-hydroxybutylidene]amino}-2-[(carboxymethyl)-C-hydroxycarbonimidoyl]ethyl]sulfanyl}-2-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-3-oxocyclopentyl]hept-5-enoate | Generator | | (5Z)-7-[(1R,2R)-5-{[(2S)-2-{[(4R)-4-amino-4-carboxy-1-hydroxybutylidene]amino}-2-[(carboxymethyl)-C-hydroxycarbonimidoyl]ethyl]sulphanyl}-2-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-3-oxocyclopentyl]hept-5-enoate | Generator | | (5Z)-7-[(1R,2R)-5-{[(2S)-2-{[(4R)-4-amino-4-carboxy-1-hydroxybutylidene]amino}-2-[(carboxymethyl)-C-hydroxycarbonimidoyl]ethyl]sulphanyl}-2-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-3-oxocyclopentyl]hept-5-enoic acid | Generator |
|
|---|
| Chemical Formula | C30H47N3O10S |
|---|
| Average Molecular Weight | 641.773 |
|---|
| Monoisotopic Molecular Weight | 641.298215429 |
|---|
| IUPAC Name | (5Z)-7-[(1R,2R)-5-{[(2S)-2-[(4R)-4-amino-4-carboxybutanamido]-2-[(carboxymethyl)carbamoyl]ethyl]sulfanyl}-2-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-3-oxocyclopentyl]hept-5-enoic acid |
|---|
| Traditional Name | (5Z)-7-[(1R,2R)-5-{[(2S)-2-[(4R)-4-amino-4-carboxybutanamido]-2-(carboxymethylcarbamoyl)ethyl]sulfanyl}-2-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-3-oxocyclopentyl]hept-5-enoic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CCCCC[C@H](O)\C=C\[C@@H]1[C@@H](C\C=C/CCCC(O)=O)C(CC1=O)SC[C@@H](NC(=O)CC[C@@H](N)C(O)=O)C(=O)NCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C30H47N3O10S/c1-2-3-6-9-19(34)12-13-20-21(10-7-4-5-8-11-27(37)38)25(16-24(20)35)44-18-23(29(41)32-17-28(39)40)33-26(36)15-14-22(31)30(42)43/h4,7,12-13,19-23,25,34H,2-3,5-6,8-11,14-18,31H2,1H3,(H,32,41)(H,33,36)(H,37,38)(H,39,40)(H,42,43)/b7-4-,13-12+/t19-,20+,21+,22+,23+,25?/m0/s1 |
|---|
| InChI Key | RJCPXHXLCWWGHO-APFRPCTBSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as oligopeptides. These are organic compounds containing a sequence of between three and ten alpha-amino acids joined by peptide bonds. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Oligopeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-oligopeptide
- Gamma-glutamyl alpha peptide
- Prostaglandin skeleton
- Eicosanoid
- Glutamine or derivatives
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- Alpha-amino acid amide
- Cysteine or derivatives
- D-alpha-amino acid
- N-substituted-alpha-amino acid
- Alpha-amino acid
- Alpha-amino acid or derivatives
- Tricarboxylic acid or derivatives
- Fatty amide
- Fatty acyl
- N-acyl-amine
- Secondary alcohol
- Secondary carboxylic acid amide
- Cyclic ketone
- Ketone
- Carboxamide group
- Amino acid or derivatives
- Amino acid
- Dialkylthioether
- Carboxylic acid
- Sulfenyl compound
- Thioether
- Primary amine
- Organic oxygen compound
- Organic nitrogen compound
- Alcohol
- Carbonyl group
- Hydrocarbon derivative
- Amine
- Organic oxide
- Organopnictogen compound
- Organosulfur compound
- Primary aliphatic amine
- Organooxygen compound
- Organonitrogen compound
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00r7-4100091000-46952d73cf49f21d796c | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05i1-0011179000-3a9d05d089ebd11bca96 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004s-3900461000-7a9b9f50ccdb2691cf24 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0avi-9301310000-088da1467bdd55576e67 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00xv-0016059000-846862813ec4ca3bccb0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01ba-0009010000-c6932bc4c2554632d9cd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0002-4903000000-3917652d16d7a5c14d59 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0avi-0015119000-b1c1a7345ce9ce89d55e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0bvj-2358879000-bf39e0dcc8c261c2fe94 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-9400200000-eab9494ac2f19dc38772 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-006x-0020009000-0eeffb6f03dfce4e2814 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0096-1930012000-a0f5bb24beb716a6f233 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9700000000-e10bf6a8bbfc904f8e17 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|