| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-10 16:59:55 UTC |
|---|
| Update Date | 2020-04-22 18:55:34 UTC |
|---|
| BMDB ID | BMDB0096022 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | UDP-N-acetyl-D-mannosamine |
|---|
| Description | UDP-N-acetyl-D-mannosamine belongs to the class of organic compounds known as pyrimidine nucleotide sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. Based on a literature review a small amount of articles have been published on UDP-N-acetyl-D-mannosamine. |
|---|
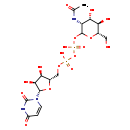
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Uridine diphosphate N-acetylgalactosamine | HMDB | | Uridine-diphosphate-N-acetylgalactosamine | HMDB | | Uridine-diphosphate-N-acetylglucosamine | HMDB | | Diphosphate N-acetylgalactosamine, uridine | MeSH, HMDB | | UDP Acetylgalactosamine | MeSH, HMDB | | Acetylgalactosamine, UDP | MeSH, HMDB | | Uridine diphosphate N acetylgalactosamine | MeSH, HMDB | | N-Acetylgalactosamine, uridine diphosphate | MeSH, HMDB |
|
|---|
| Chemical Formula | C17H27N3O17P2 |
|---|
| Average Molecular Weight | 607.3537 |
|---|
| Monoisotopic Molecular Weight | 607.081569477 |
|---|
| IUPAC Name | [({[(2S,3R,4S,5S)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(3R,4S,5R,6S)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy})phosphinic acid |
|---|
| Traditional Name | {[(2S,3R,4S,5S)-5-(2,4-dioxo-3H-pyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy([(3R,4S,5R,6S)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy)phosphinic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CC(=O)N[C@@H]1[C@H](O)[C@@H](O)[C@H](CO)OC1OP(O)(=O)OP(O)(=O)OC[C@@H]1O[C@@H]([C@@H](O)[C@H]1O)N1C=CC(=O)NC1=O |
|---|
| InChI Identifier | InChI=1S/C17H27N3O17P2/c1-6(22)18-10-13(26)11(24)7(4-21)35-16(10)36-39(31,32)37-38(29,30)33-5-8-12(25)14(27)15(34-8)20-3-2-9(23)19-17(20)28/h2-3,7-8,10-16,21,24-27H,4-5H2,1H3,(H,18,22)(H,29,30)(H,31,32)(H,19,23,28)/t7-,8-,10+,11-,12-,13-,14-,15-,16?/m0/s1 |
|---|
| InChI Key | LFTYTUAZOPRMMI-IUPDHGCPSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine nucleotide sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine nucleotide sugars |
|---|
| Direct Parent | Pyrimidine nucleotide sugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine nucleotide sugar
- Pyrimidine ribonucleoside diphosphate
- Pentose phosphate
- Pentose-5-phosphate
- N-acyl-alpha-hexosamine
- Glycosyl compound
- N-glycosyl compound
- Monosaccharide phosphate
- Organic pyrophosphate
- Pyrimidone
- Monoalkyl phosphate
- Hydropyrimidine
- Monosaccharide
- Organic phosphoric acid derivative
- Oxane
- Phosphoric acid ester
- Pyrimidine
- Alkyl phosphate
- Vinylogous amide
- Acetamide
- Tetrahydrofuran
- Heteroaromatic compound
- Urea
- Secondary carboxylic acid amide
- Secondary alcohol
- Carboxamide group
- Lactam
- Azacycle
- Carboxylic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Organopnictogen compound
- Organooxygen compound
- Alcohol
- Primary alcohol
- Organic nitrogen compound
- Organic oxygen compound
- Organic oxide
- Carbonyl group
- Hydrocarbon derivative
- Organonitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0f96-5924660000-c11cc7fa21a050c0f373 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0h03-7952838000-56b15bfcc0138d466a26 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0910020000-4282200321d0bfe45943 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-3921000000-6076288d599ab3313479 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-5910000000-14d31d6720f9ec539675 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-06ri-9502331000-138ea2dd91899208164e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03dl-9504010000-0424c67ad101c9f6be36 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-06r6-5901000000-dce542e7a9e33e0e747c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000029000-6a481f04b19fa3177889 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a6s-5100192000-07c7ec84c4bab85e334a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-053u-4719110000-3a1b07828c441361af2f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00mk-0000982000-8e4f05f581cea906b6bb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0hi2-0213591000-da4a947496986866c59e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4j-6980020000-82befaa44d12d4c3c15d | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|