| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-10 17:02:38 UTC |
|---|
| Update Date | 2020-05-11 20:27:40 UTC |
|---|
| BMDB ID | BMDB0096188 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
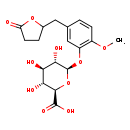
| Common Name | 5-(3',4'-Dihydroxyphenyl)-gamma-valerolactone-4'-O-methyl-3'-O-glucuronide |
|---|
| Description | 5-(3',4'-Dihydroxyphenyl)-gamma-valerolactone-4'-O-methyl-3'-O-glucuronide belongs to the class of organic compounds known as phenolic glycosides. These are organic compounds containing a phenolic structure attached to a glycosyl moiety. Some examples of phenolic structures include lignans, and flavonoids. Among the sugar units found in natural glycosides are D-glucose, L-Fructose, and L rhamnose. 5-(3',4'-Dihydroxyphenyl)-gamma-valerolactone-4'-O-methyl-3'-O-glucuronide is an extremely weak basic (essentially neutral) compound (based on its pKa). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 5-(3',4'-Dihydroxyphenyl)-g-valerolactone-4'-O-methyl-3'-O-glucuronide | Generator | | 5-(3',4'-Dihydroxyphenyl)-γ-valerolactone-4'-O-methyl-3'-O-glucuronide | Generator | | (2R,3R,4R,5S,6R)-3,4,5-Trihydroxy-6-{2-methoxy-5-[(5-oxooxolan-2-yl)methyl]phenoxy}oxane-2-carboxylate | Generator |
|
|---|
| Chemical Formula | C18H22O10 |
|---|
| Average Molecular Weight | 398.3613 |
|---|
| Monoisotopic Molecular Weight | 398.121296924 |
|---|
| IUPAC Name | (2R,3R,4R,5S,6R)-3,4,5-trihydroxy-6-{2-methoxy-5-[(5-oxooxolan-2-yl)methyl]phenoxy}oxane-2-carboxylic acid |
|---|
| Traditional Name | (2R,3R,4R,5S,6R)-3,4,5-trihydroxy-6-{2-methoxy-5-[(5-oxooxolan-2-yl)methyl]phenoxy}oxane-2-carboxylic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | COC1=C(O[C@H]2O[C@H]([C@H](O)[C@@H](O)[C@@H]2O)C(O)=O)C=C(CC2CCC(=O)O2)C=C1 |
|---|
| InChI Identifier | InChI=1S/C18H22O10/c1-25-10-4-2-8(6-9-3-5-12(19)26-9)7-11(10)27-18-15(22)13(20)14(21)16(28-18)17(23)24/h2,4,7,9,13-16,18,20-22H,3,5-6H2,1H3,(H,23,24)/t9?,13-,14-,15+,16-,18+/m1/s1 |
|---|
| InChI Key | VZGJAVFCUBLIJF-ZOQSEFCISA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phenolic glycosides. These are organic compounds containing a phenolic structure attached to a glycosyl moiety. Some examples of phenolic structures include lignans, and flavonoids. Among the sugar units found in natural glycosides are D-glucose, L-Fructose, and L rhamnose. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Phenolic glycosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Phenolic glycoside
- 1-o-glucuronide
- O-glucuronide
- Glucuronic acid or derivatives
- O-glycosyl compound
- Phenoxy compound
- Anisole
- Methoxybenzene
- Phenol ether
- Alkyl aryl ether
- Beta-hydroxy acid
- Hydroxy acid
- Gamma butyrolactone
- Pyran
- Oxane
- Dicarboxylic acid or derivatives
- Benzenoid
- Monosaccharide
- Monocyclic benzene moiety
- Tetrahydrofuran
- Carboxylic acid ester
- Secondary alcohol
- Lactone
- Carboxylic acid
- Acetal
- Carboxylic acid derivative
- Organoheterocyclic compound
- Oxacycle
- Ether
- Polyol
- Alcohol
- Carbonyl group
- Hydrocarbon derivative
- Organic oxide
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-057i-9168000000-4b240e84031837b7c2d0 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-00di-4010039000-9cb225a0401b5d7eb10f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_3_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_4_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("5-(3',4'-Dihydroxyphenyl)-gamma-valerolactone-4'-O-methyl-3'-O-glucuronide,3TMS,#3" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00ea-0269000000-4fd7a2f24739bfa567ca | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-0941000000-dd9e653a9c7150ae3ede | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00dr-1920000000-dee7df14ecea5c56a020 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0fdk-1339000000-8c0823ec188d0ec26ad8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00fr-3595000000-bab6b3a29126785ae972 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9430000000-c444629511c26892638a | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|