| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:27:10 UTC |
|---|
| Update Date | 2020-06-04 20:45:42 UTC |
|---|
| BMDB ID | BMDB0000235 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Thiamine |
|---|
| Description | Thiamine, also known as vitamin B1 or aneurin, belongs to the class of organic compounds known as thiamines. Thiamines are compounds containing a thiamine moiety, which is structurally characterized by a 3-[(4-Amino-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-thiazol-5-yl backbone. Thiamine is a drug which is used for the treatment of thiamine and niacin deficiency states, korsakov's alcoholic psychosis, wernicke-korsakov syndrome, delirium, and peripheral neuritis. Thiamine exists as a solid, possibly soluble (in water), and a very strong basic compound (based on its pKa) molecule. Thiamine exists in all living species, ranging from bacteria to humans. In cattle, thiamine is involved in the metabolic pathway called the valine, leucine, and isoleucine degradation pathway. Thiamine is a potentially toxic compound. |
|---|
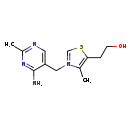
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-(4-AMINO-2-methyl-pyrimidin-5-ylmethyl)-5-(2-hydroxy-ethyl)-4-methyl-thiazol-3-ium | ChEBI | | Aneurin | ChEBI | | Antiberiberi factor | ChEBI | | Thiamin | ChEBI | | Thiamine(1+) ion | ChEBI | | Thiaminium | ChEBI | | Vitamin b1 | ChEBI | | Apate drops | HMDB | | Beatine | HMDB | | Bedome | HMDB | | Begiolan | HMDB | | Benerva | HMDB | | Bequin | HMDB | | Berin | HMDB | | Betalin S | HMDB | | Betaxin | HMDB | | Bethiazine | HMDB | | Beuion | HMDB | | Bevitex | HMDB | | Bevitine | HMDB | | Bewon | HMDB | | Biamine | HMDB | | Bithiamin | HMDB | | Biuno | HMDB | | Bivatin | HMDB | | Bivita | HMDB | | Cernevit-12 | HMDB | | Clotiamina | HMDB | | Eskapen | HMDB | | Eskaphen | HMDB | | Hybee | HMDB | | Lixa-beta | HMDB | | Metabolin | HMDB | | Slowten | HMDB | | THD | HMDB | | Thiadoxine | HMDB | | Thiaminal | HMDB | | Thiamol | HMDB | | Thiavit | HMDB | | Tiamidon | HMDB | | Tiaminal | HMDB | | Trophite | HMDB | | Vetalin S | HMDB | | VIB | HMDB | | Vinothiam | HMDB | | Vitaneuron | HMDB | | Mononitrate, thiamine | MeSH, HMDB | | Thiamine mononitrate | MeSH, HMDB | | Vitamin b 1 | MeSH, HMDB |
|
|---|
| Chemical Formula | C12H17N4OS |
|---|
| Average Molecular Weight | 265.355 |
|---|
| Monoisotopic Molecular Weight | 265.112306876 |
|---|
| IUPAC Name | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-hydroxyethyl)-4-methyl-1,3-thiazol-3-ium |
|---|
| Traditional Name | thiamine |
|---|
| CAS Registry Number | 59-43-8 |
|---|
| SMILES | CC1=C(CCO)SC=[N+]1CC1=CN=C(C)N=C1N |
|---|
| InChI Identifier | InChI=1S/C12H17N4OS/c1-8-11(3-4-17)18-7-16(8)6-10-5-14-9(2)15-12(10)13/h5,7,17H,3-4,6H2,1-2H3,(H2,13,14,15)/q+1 |
|---|
| InChI Key | JZRWCGZRTZMZEH-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as thiamines. Thiamines are compounds containing a thiamine moiety, which is structurally characterized by a 3-[(4-Amino-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-thiazol-5-yl backbone. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Diazines |
|---|
| Sub Class | Pyrimidines and pyrimidine derivatives |
|---|
| Direct Parent | Thiamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Thiamine
- 4,5-disubstituted 1,3-thiazole
- Aminopyrimidine
- Imidolactam
- Azole
- Thiazole
- Heteroaromatic compound
- Azacycle
- Alcohol
- Organopnictogen compound
- Primary amine
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Organic oxygen compound
- Hydrocarbon derivative
- Amine
- Organic cation
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected and Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Membrane
- Mitochondria
- Myelin sheath
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 248 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 500.0 mg/mL | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00e9-3890000000-9f8b525433ab279ad512 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-9442000000-05ab93d3afb7b6f538e9 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, N/A (Annotated) | splash10-000i-0109000000-48864f31ba475645654b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, N/A (Annotated) | splash10-05o1-9200000000-2ec0f3e9e364cffde16e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, N/A (Annotated) | splash10-0aou-9000000000-0184506492d9f5b68b97 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-03di-0090000000-872150be83cbd75d8ecf | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0002-0930000000-ecf71c75bdea859139d5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0002-0900000000-4d9113fdc633fb1ccbb5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0002-0900000000-4a8016299623af0c3027 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0002-1900000000-773912c9f325c419c77d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-014i-0390000000-95f3ca57c95542bdc7e7 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-00di-0900000000-e079d1fc1e06396dc524 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-00di-0900000000-ebb16d4f204f291b9a3a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-00e9-4900000000-ad4d4779d76b9edb7ec5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-001i-9600000000-b631eed5be831527fc17 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-00di-0900000000-50f2f613f576814eb627 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-002b-0900000000-2f184bbfec6bc0ef00c6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-001i-9000000000-35bc744495a2570a565f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-00di-1900000000-d8a7ac327273c32b4524 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0002-0920000000-ca80ef6513c61be5b5e9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-03di-0090000000-872150be83cbd75d8ecf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0190000000-183af2449d1439af9825 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-0090000000-2230ebc7c6b73192c26f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-7920000000-75670df9647a475ca92c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-3090000000-d0ff9a733629a86b75c7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03dj-3690000000-db3f204af490d7e1722e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0005-9000000000-5196f2b4daab995967bf | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| General References | - Agostini TS, Scherer R, Godoy HT: Simultaneous determination of B-group vitamins in enriched Brazilian dairy products. Crit Rev Food Sci Nutr. 2007;47(5):435-9. doi: 10.1080/10408390600846309. [PubMed:17558655 ]
- Gaucheron F: Milk and dairy products: a unique micronutrient combination. J Am Coll Nutr. 2011 Oct;30(5 Suppl 1):400S-9S. [PubMed:22081685 ]
- Park, Y. W; Juárez, Manuela ; Ramos, M.; Haenlein, G. F. W. (2007). Park, Y. W; Juárez, Manuela ; Ramos, M.; Haenlein, G. F. W.. Physico-chemical characteristics of goat and sheep milk. Small Ruminant Res.(2007) 68:88-113 doi: 10.1016/j.smallrumres.2006.09.013. Small Ruminant Research.
- A. Foroutan et al. (2019). A. Foroutan et al. The Chemical Composition of Commercial Cow's Milk (in preparation). Journal of Agricultural and Food Chemistry.
- USDA Food Composition Databases [Link]
|
|---|