| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:27:16 UTC |
|---|
| Update Date | 2020-05-20 23:10:48 UTC |
|---|
| BMDB ID | BMDB0000241 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Protoporphyrin IX |
|---|
| Description | Protoporphyrin IX, also known as porphyrinogen IX or H2PPIX, belongs to the class of organic compounds known as porphyrins. Porphyrins are compounds containing a fundamental skeleton of four pyrrole nuclei united through the alpha-positions by four methine groups to form a macrocyclic structure. Protoporphyrin IX exists in all living species, ranging from bacteria to plants to humans. Based on a literature review a significant number of articles have been published on Protoporphyrin IX. |
|---|
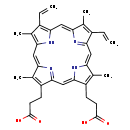
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3,3'-(3,7,12,17-Tetramethyl-8,13-divinyl-21H,23H-porphine-2,18-diyl)-bis-propionic acid | ChEBI | | 3,7,12,17-Tetramethyl-8,13-divinylporphyrin-2,18-dipropanoic acid | ChEBI | | H2PpIX | ChEBI | | Kammerer's prophyrin | ChEBI | | Ooporphyrin | ChEBI | | Porphyrinogen IX | ChEBI | | 3,3'-(3,7,12,17-Tetramethyl-8,13-divinyl-21H,23H-porphine-2,18-diyl)-bis-propionate | Generator | | 3,7,12,17-Tetramethyl-8,13-divinylporphyrin-2,18-dipropanoate | Generator | | 3,3'-(3,7,12,17-Tetramethyl-8,13-divinylporphine-2,18-diyl)di | HMDB | | PpIX | HMDB | | Protoporphyrin | HMDB | | Protoporphyrin-"ix" | HMDB | | Protoporphyrin-IX | HMDB |
|
|---|
| Chemical Formula | C34H34N4O4 |
|---|
| Average Molecular Weight | 562.6582 |
|---|
| Monoisotopic Molecular Weight | 562.258005596 |
|---|
| IUPAC Name | 3-[20-(2-carboxyethyl)-9,14-diethenyl-5,10,15,19-tetramethyl-21,22,23,24-tetraazapentacyclo[16.2.1.1^{3,6}.1^{8,11}.1^{13,16}]tetracosa-1(21),2,4,6,8(23),9,11,13,15,17,19-undecaen-4-yl]propanoic acid |
|---|
| Traditional Name | 3-[20-(2-carboxyethyl)-9,14-diethenyl-5,10,15,19-tetramethyl-21,22,23,24-tetraazapentacyclo[16.2.1.1^{3,6}.1^{8,11}.1^{13,16}]tetracosa-1(21),2,4,6,8(23),9,11,13,15,17,19-undecaen-4-yl]propanoic acid |
|---|
| CAS Registry Number | 553-12-8 |
|---|
| SMILES | CC1=C(CCC(O)=O)/C2=C/C3=N/C(=C\C4=C(C)C(C=C)=C(N4)/C=C4\N=C(\C=C\1/N\2)C(C=C)=C4C)/C(C)=C3CCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C34H34N4O4/c1-7-21-17(3)25-13-26-19(5)23(9-11-33(39)40)31(37-26)16-32-24(10-12-34(41)42)20(6)28(38-32)15-30-22(8-2)18(4)27(36-30)14-29(21)35-25/h7-8,13-16,35,38H,1-2,9-12H2,3-6H3,(H,39,40)(H,41,42)/b25-13-,26-13-,27-14-,28-15-,29-14-,30-15-,31-16-,32-16- |
|---|
| InChI Key | KSFOVUSSGSKXFI-UJJXFSCMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as porphyrins. Porphyrins are compounds containing a fundamental skeleton of four pyrrole nuclei united through the alpha-positions by four methine groups to form a macrocyclic structure. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Tetrapyrroles and derivatives |
|---|
| Sub Class | Porphyrins |
|---|
| Direct Parent | Porphyrins |
|---|
| Alternative Parents | Not Available |
|---|
| Substituents | Not Available |
|---|
| Molecular Framework | Not Available |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Cytoplasm
- Membrane
- Mitochondria
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 300 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 0.169 mg/mL at 25 °C | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0uxu-1000290000-e22236fe3cca8eed1ecf | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-6000094000-b9c12952e100c5144cab | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Protoporphyrin IX,1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0000090000-5d5be53410134c5719ab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kb-1000590000-2d4157b9f73534d95a9c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-1000910000-07524a9291a1921dfafc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0000090000-5df26c1cfb9a0b16a91a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0296-1000190000-2d4bcb49201fc10adc09 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000230000-064fb944a966d08c06bf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0000090000-87c1740ecd1a28f4a2d9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03xu-0000590000-99160b987bb7e62ab362 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-0000900000-f2d9118441a9c8c47d79 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dj-0000090000-d428ac6dc2fc4794cc41 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0j4j-0000090000-2b065966ec57bb97d511 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uk9-1000970000-b435817e4dc349e65ab3 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|