| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:32:02 UTC |
|---|
| Update Date | 2020-05-11 20:00:42 UTC |
|---|
| BMDB ID | BMDB0000518 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Chenodeoxycholic acid |
|---|
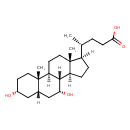
| Description | Chenodeoxycholic acid, also known as anthropodeoxycholate or chenix, belongs to the class of organic compounds known as dihydroxy bile acids, alcohols and derivatives. Dihydroxy bile acids, alcohols and derivatives are compounds containing or derived from a bile acid or alcohol, and which bears exactly two carboxylic acid groups. Chenodeoxycholic acid is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3alpha,7alpha-Dihydroxy-5beta-cholanic acid | ChEBI | | 7alpha-Hydroxylithocholic acid | ChEBI | | Anthropodeoxycholic acid | ChEBI | | Anthropodesoxycholic acid | ChEBI | | CDCA | ChEBI | | Chenic acid | ChEBI | | Chenix | ChEBI | | Chenodiol | ChEBI | | Gallodesoxycholic acid | ChEBI | | 3a,7a-Dihydroxy-5b-cholanate | Generator | | 3a,7a-Dihydroxy-5b-cholanic acid | Generator | | 3alpha,7alpha-Dihydroxy-5beta-cholanate | Generator | | 3Α,7α-dihydroxy-5β-cholanate | Generator | | 3Α,7α-dihydroxy-5β-cholanic acid | Generator | | 7a-Hydroxylithocholate | Generator | | 7a-Hydroxylithocholic acid | Generator | | 7alpha-Hydroxylithocholate | Generator | | 7Α-hydroxylithocholate | Generator | | 7Α-hydroxylithocholic acid | Generator | | Anthropodeoxycholate | Generator | | Anthropodesoxycholate | Generator | | Chenate | Generator | | Gallodesoxycholate | Generator | | Chenodeoxycholate | Generator | | (+)-Chenodeoxycholate | HMDB | | (+)-Chenodeoxycholic acid | HMDB | | (3a,5b,7a)-3,7-Dihydroxy-cholan-24-Oate | HMDB | | (3a,5b,7a)-3,7-Dihydroxy-cholan-24-Oic acid | HMDB | | 3a,7a-Dihydroxy-5b,14a,17b-cholanate | HMDB | | 3a,7a-Dihydroxy-5b,14a,17b-cholanic acid | HMDB | | 3a,7a-Dihydroxy-5b-cholan-24-Oate | HMDB | | 3a,7a-Dihydroxy-5b-cholan-24-Oic acid | HMDB | | 7a-Hydroxy-desoxycholsaeure | HMDB | | Chenodesoxycholsaeure | HMDB | | Acid, chenique | HMDB | | Chenofalk | HMDB | | Chenophalk | HMDB | | Acid, chenodeoxycholic | HMDB | | Chenodeoxycholate, sodium | HMDB | | Quenocol | HMDB | | Solvay brand OF chenodeoxycholic acid | HMDB | | Antigen brand OF chenodeoxycholic acid | HMDB | | Falk brand OF chenodeoxycholic acid | HMDB | | Quenobilan | HMDB | | Sodium chenodeoxycholate | HMDB | | Tramedico brand OF chenodeoxycholic acid | HMDB | | Zambon brand OF chenodeoxycholic acid | HMDB | | Acid, chenic | HMDB | | Acid, gallodesoxycholic | HMDB | | Chenique acid | HMDB | | Estedi brand OF chenodeoxycholic acid | HMDB | | Henohol | HMDB |
|
|---|
| Chemical Formula | C24H40O4 |
|---|
| Average Molecular Weight | 392.572 |
|---|
| Monoisotopic Molecular Weight | 392.292659768 |
|---|
| IUPAC Name | (4R)-4-[(1S,2S,5R,7S,9R,10R,11S,14R,15R)-5,9-dihydroxy-2,15-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanoic acid |
|---|
| Traditional Name | (4R)-4-[(1S,2S,5R,7S,9R,10R,11S,14R,15R)-5,9-dihydroxy-2,15-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanoic acid |
|---|
| CAS Registry Number | 474-25-9 |
|---|
| SMILES | [H][C@@]1(CC[C@@]2([H])[C@]3([H])[C@H](O)C[C@]4([H])C[C@H](O)CC[C@]4(C)[C@@]3([H])CC[C@]12C)[C@H](C)CCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C24H40O4/c1-14(4-7-21(27)28)17-5-6-18-22-19(9-11-24(17,18)3)23(2)10-8-16(25)12-15(23)13-20(22)26/h14-20,22,25-26H,4-13H2,1-3H3,(H,27,28)/t14-,15+,16-,17-,18+,19+,20-,22+,23+,24-/m1/s1 |
|---|
| InChI Key | RUDATBOHQWOJDD-BSWAIDMHSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dihydroxy bile acids, alcohols and derivatives. Dihydroxy bile acids, alcohols and derivatives are compounds containing or derived from a bile acid or alcohol, and which bears exactly two carboxylic acid groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Dihydroxy bile acids, alcohols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Dihydroxy bile acid, alcohol, or derivatives
- 3-hydroxysteroid
- Hydroxysteroid
- 3-alpha-hydroxysteroid
- 7-hydroxysteroid
- Cyclic alcohol
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organooxygen compound
- Alcohol
- Organic oxygen compound
- Carbonyl group
- Hydrocarbon derivative
- Organic oxide
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 165 - 167 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 0.0899 mg/mL | Not Available | | LogP | 4.15 | SANGSTER (1993) |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (3 TMS) | splash10-0a6u-3920000000-ce93b24c6e2568b6087d | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-0a6u-3920000000-ce93b24c6e2568b6087d | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01rt-0419000000-6a92f910581240163a99 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0006-1110390000-7a6c62344d1371b74081 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_3_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, N/A (Annotated) | splash10-0a4i-0009000000-82ed6dc62b49ac0340e6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, N/A (Annotated) | splash10-01pa-2930000000-64edc819ad56b842cdba | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, N/A (Annotated) | splash10-053r-6900000000-c97325e1ca9bcff75af1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , negative | splash10-0002-0009000000-ad2d2440db7f3f1c8a2e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0006-0009000000-2df562379077bf0f4b2a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0006-0009000000-ff13f6ad8405e3969b6c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0006-0009000000-1af3ba6ee17285da06a2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0009000000-5409557fb9a76a8a8e74 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0006-0009000000-c77d97d0dfe48497b263 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-0006-0009000000-d38671ccaaa16ac6d1f1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0596-0529000000-1ed436cc96fa78f90b88 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0009000000-25ba2afe78d7d8070765 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0006-0019000000-c31d129de7de9df05167 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0006-0009000000-fc161813afdb6d319d8d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0006-0009000000-ee319e0cbd9987920deb | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0009000000-858f2baceacfba406468 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-056r-0009000000-997e61e986e67265241c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-056r-0009000000-81134947694f847d1c65 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-02t9-1219000000-6a0ddebebacac090c27e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0009000000-efdaad69eee0ae934dd5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-006x-1009000000-15b4edab9d05e4c3693a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9006000000-9291e69db3a3c47ecd97 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0009000000-7631731446db77938069 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-006x-0009000000-39e7e555dc6b36ada2af | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000i-1009000000-351c21e3dab693818b22 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|