| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:36:00 UTC |
|---|
| Update Date | 2020-03-13 16:23:35 UTC |
|---|
| BMDB ID | BMDB0000751 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | L-Threo-2-pentulose |
|---|
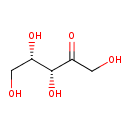
| Description | D-Xylulose, also known as L-lyxulose or L-threo-pentulose, belongs to the class of organic compounds known as pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. D-Xylulose is an extremely weak basic (essentially neutral) compound (based on its pKa). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| L-Lyxulose | ChEBI | | L-Threo-pent-2-ulose | ChEBI | | L-Threo-pentulose | ChEBI | | L-Xul | ChEBI | | L-Xylulose | HMDB |
|
|---|
| Chemical Formula | C5H10O5 |
|---|
| Average Molecular Weight | 150.1299 |
|---|
| Monoisotopic Molecular Weight | 150.05282343 |
|---|
| IUPAC Name | (3R,4S)-1,3,4,5-tetrahydroxypentan-2-one |
|---|
| Traditional Name | xylulose |
|---|
| CAS Registry Number | 527-50-4 |
|---|
| SMILES | OC[C@H](O)[C@@H](O)C(=O)CO |
|---|
| InChI Identifier | InChI=1S/C5H10O5/c6-1-3(8)5(10)4(9)2-7/h3,5-8,10H,1-2H2/t3-,5+/m0/s1 |
|---|
| InChI Key | ZAQJHHRNXZUBTE-WVZVXSGGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Pentoses |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pentose monosaccharide
- Beta-hydroxy ketone
- Acyloin
- Alpha-hydroxy ketone
- Secondary alcohol
- Ketone
- Polyol
- Organic oxide
- Hydrocarbon derivative
- Primary alcohol
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | Not Available |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03di-9000000000-5e0dfc74544a16cb6a2b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-00g3-9127300000-b6aed5448276d0184bc2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-2900000000-3e1ebc07944b3d190ec8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-9300000000-bca65135a1359f774229 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08fv-9000000000-71ba0bd1388ac4ba066d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000j-9400000000-a8afb4fe5a51e50f0c41 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-052r-9200000000-bbc5e84222a57470dc89 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-ce9ee9286404da2668c5 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|